2KVO
 
 | | Solution NMR structure of Photosystem II reaction center Psb28 protein from Synechocystis sp.(strain PCC 6803), Northeast Structural Genomics Consortium Target SgR171 | | Descriptor: | Photosystem II reaction center psb28 protein | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Wang, D, Ciccosanti, C, Hamilton, K, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-22 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of photosystem II reaction center protein Psb28 from Synechocystis sp. Strain PCC 6803.
Proteins, 79, 2011
|
|
2LK2
 
 | | Solution NMR structure of homeobox domain (171-248) of human homeobox protein TGIF1, Northeast Structural Genomics Consortium Target HR4411B | | Descriptor: | Homeobox protein TGIF1 | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Shastry, R, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-10-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of homeobox domain (171-248) of human homeobox protein TGIF1, Northeast Structural Genomics Consortium Target HR4411B.
To be Published
|
|
3LMO
 
 | | Crystal Structure of specialized acyl carrier protein (RPA2022) from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR324 | | Descriptor: | Specialized acyl carrier protein | | Authors: | Forouhar, F, Rossi, P, Lew, S, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long-chain fatty acids in open and closed conformations.
Biochemistry, 51, 2012
|
|
1TVG
 
 | | X-ray structure of human PP25 gene product, HSPC034. Northeast Structural Genomics Target HR1958. | | Descriptor: | CALCIUM ION, LOC51668 protein, SAMARIUM (III) ION | | Authors: | Kuzin, A.P, Vorobiev, S.M, Lee, I, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
2KW2
 
 | | Solution NMR of the specialized acyl carrier protein (RPA2022) from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR324 | | Descriptor: | Specialized acyl carrier protein | | Authors: | Rossi, P, Lee, H, Wohlbold, T.J, Valafar, H, Lemak, A, Ertekin, A, Forouhar, F, Ciccosanti, C, Rost, B, Acton, T, Xiao, R, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-30 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long-chain fatty acids in open and closed conformations.
Biochemistry, 51, 2012
|
|
5ZKL
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT12 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
2AYJ
 
 | | Solution structure of 50S ribosomal protein L40e from Sulfolobus solfataricus | | Descriptor: | 50S ribosomal protein L40e, ZINC ION | | Authors: | Wu, B, Yee, A, Lukin, J, Lemak, A, Semesi, A, Ramelot, T, Kennedy, M, Edward, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-07 | | Release date: | 2006-08-22 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ribosomal protein L40E, a unique C4 zinc finger protein encoded by archaeon Sulfolobus solfataricus
Protein Sci., 17, 2008
|
|
7K3H
 
 | | Crystal structure of deep network hallucinated protein 0217 | | Descriptor: | Network hallucinated protein 0217 | | Authors: | Pellock, S.J, Anishchenko, I, Chidyausiku, T.M, Bera, A.K, DiMaio, F, Baker, D. | | Deposit date: | 2020-09-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo protein design by deep network hallucination.
Nature, 600, 2021
|
|
8RD6
 
 | |
5ZKM
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA TCTTCC | | Descriptor: | DNA (5'-D(P*TP*CP*TP*TP*CP*C)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
3H9X
 
 | | Crystal Structure of the PSPTO_3016 protein from Pseudomonas syringae, Northeast Structural Genomics Consortium Target PsR293 | | Descriptor: | uncharacterized protein PSPTO_3016 | | Authors: | Seetharaman, J, Lew, S, Forouhar, F, Hamilton, H, Xiao, R, Ciccosanti, C, Foote, E.L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Solution NMR and X-ray crystal structures of Pseudomonas syringae Pspto_3016 from protein domain family PF04237 (DUF419) adopt a "double wing" DNA binding motif.
J.Struct.Funct.Genom., 13, 2012
|
|
6JIQ
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6JIP
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), PENTAETHYLENE GLYCOL, SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
4FPW
 
 | | Crystal Structure of CalU16 from Micromonospora echinospora. Northeast Structural Genomics Consortium Target MiR12. | | Descriptor: | CalU16 | | Authors: | Seetharaman, J, Lew, S, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Phillips Jr, G.N, Kennedy, M.A, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-22 | | Release date: | 2012-12-12 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
1ZCE
 
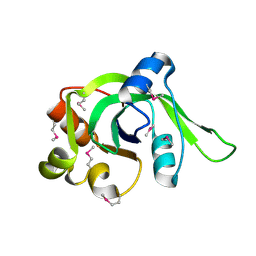 | | X-Ray Crystal Structure of Protein Atu2648 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR33. | | Descriptor: | hypothetical protein Atu2648 | | Authors: | Forouhar, F, Chen, Y, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-11 | | Release date: | 2005-04-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
4J0B
 
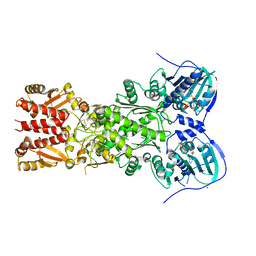 | | Structure of mitochondrial Hsp90 (TRAP1) with ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, COBALT (II) ION, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-30 | | Release date: | 2014-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
4IVG
 
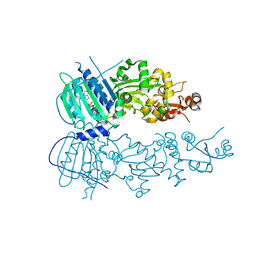 | | Crystal structure of mitochondrial Hsp90 (TRAP1) NTD-Middle domain dimer with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-22 | | Release date: | 2014-01-22 | | Last modified: | 2014-08-27 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
4IYN
 
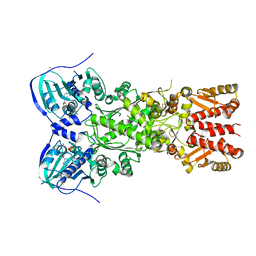 | | Structure of mitochondrial Hsp90 (TRAP1) with ADP-ALF4- | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
4IPE
 
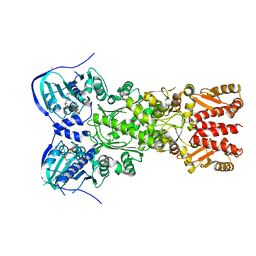 | | Crystal structure of mitochondrial Hsp90 (TRAP1) with AMPPNP | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2014-08-27 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
1R57
 
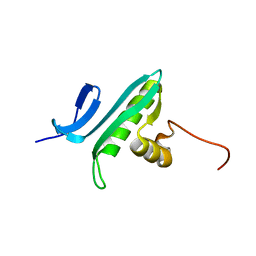 | | NMR Solution Structure of a GCN5-like putative N-acetyltransferase from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR31 | | Descriptor: | conserved hypothetical protein | | Authors: | Cort, J.R, Acton, T.B, Ma, L, Xiao, R.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-10-09 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an acetyl-CoA binding protein from Staphylococcus aureus representing a novel subfamily of GCN5-related N-acetyltransferase-like proteins.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
7UCP
 
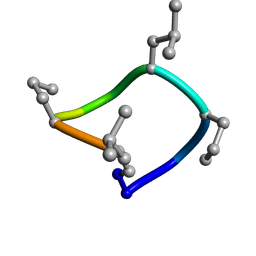 | | computationally designed macrocycle | | Descriptor: | computationally designed cyclic peptide D8.3.p2 | | Authors: | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | Deposit date: | 2022-03-17 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
7M0Q
 
 | |
2EVE
 
 | | X-Ray Crystal Structure of Protein PSPTO5229 from Pseudomonas syringae. Northeast Structural Genomics Consortium Target PsR62 | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ... | | Authors: | Forouhar, F, Zhou, W, Belachew, A, Jayaraman, S, Ciao, M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
2G2X
 
 | | X-Ray Crystal Structure Protein Q88CH6 from Pseudomonas putida. Northeast Structural Genomics Consortium Target PpR72. | | Descriptor: | SULFATE ION, hypothetical protein PP5205 | | Authors: | Kuzin, A.P, Chen, Y, Abashidze, M, Acton, T, Conover, K, Janjua, H, Ma, L.-C, Ho, C.K, Cunningham, K, Montelione, G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-02-16 | | Release date: | 2006-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
2JS7
 
 | | Solution NMR structure of human myeloid differentiation primary response (MyD88). Northeast Structural Genomics target HR2869A | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Rossi, P, Ramelot, T.A, Tao, X, Ciano, M, Ho, C, Ma, L.-C, Xiao, R, Acton, T.B, Kennedy, M.A, Tong, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-10-23 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Human Myeloid Differentiation Primary Response (MyD88).
To be Published
|
|
