8CTO
 
 | |
8CUN
 
 | |
8CWA
 
 | |
7M5T
 
 | |
2RN7
 
 | | NMR solution structure of TnpE protein from Shigella flexneri. Northeast Structural Genomics Target SfR125 | | Descriptor: | IS629 orfA | | Authors: | Ramelot, T.A, Cort, J.R, Semesi, A, Garcia, M, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-08 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of TnpE protein from Shigella flexneri. Northeast
Structural Genomics Target SfR125
To be Published
|
|
1JJG
 
 | | Solution Structure of Myxoma Virus Protein M156R | | Descriptor: | M156R | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-07-05 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2alpha.
J.Mol.Biol., 322, 2002
|
|
2KHD
 
 | | Solution NMR structure of VC_A0919 from Vibrio cholerae. Northeast Structural Genomics Consortium Target VcR52 | | Descriptor: | Uncharacterized protein VC_A0919 | | Authors: | Ramelot, T.A, Cort, J.R, Wang, H, Ciccosanti, C, Jiang, M, Liu, J, Rost, B, Swapna, G.V.T, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-02 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of VC_A0919 from Vibrio cholerae. Northeast Structural Genomics Consortium Target VcR52.
To be Published
|
|
1XPW
 
 | | Solution NMR Structure of human protein HSPCO34. Northeast Structural Genomics Target HR1958 | | Descriptor: | LOC51668 protein | | Authors: | Ramelot, T.A, Xiao, R, Ma, L.C, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
1Q48
 
 | | Solution NMR Structure of The Haemophilus Influenzae Iron-Sulfur Cluster Assembly Protein U (IscU) with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target IR24. This protein is not apo, it is a model without zinc binding constraints. | | Descriptor: | NifU-like protein | | Authors: | Ramelot, T.A, Cort, J.R, Xiao, R, Shastry, R, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site.
J.Mol.Biol., 344, 2004
|
|
2KEN
 
 | | Solution NMR structure of the OB domain (residues 67-166) of MM0293 from Methanosarcina mazei. Northeast Structural Genomics Consortium target MaR214a. | | Descriptor: | Conserved protein | | Authors: | Ramelot, T.A, Ding, K, Magliqui, M, Jiang, M, Ciccosanti, C, Xiao, R, Lui, J, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the OB domain of MM0293 from the archea Methanosarcina mazei
To be Published
|
|
2K75
 
 | | Solution NMR structure of the OB domain of Ta0387 from Thermoplasma acidophilum. Northeast Structural Genomics Consortium target TaR80b. | | Descriptor: | uncharacterized protein Ta0387 | | Authors: | Ramelot, T.A, Ding, K, Lee, D, Jiang, M, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-01 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the OB domain of Ta0387 from Thermoplasma acidophilum.
To be Published
|
|
2KKP
 
 | | Solution NMR structure of the phage integrase SAM-like Domain from Moth 1796 from Moorella thermoacetica. Northeast Structural Genomics Consortium Target MtR39K (residues 64-171). | | Descriptor: | Phage integrase | | Authors: | Ramelot, T.A, Wang, H, Ciccosanti, C, Jang, M, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the phage integrase SAM-like Domain from Moth 1796
from Moorella thermoacetica. Northeast
Structural Genomics Consortium Target MtR39K (residues 64-171).
To be Published
|
|
2K9Q
 
 | | Solution NMR structure of HTH_XRE family transcriptional regulator BT_p548217 from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR244. | | Descriptor: | uncharacterized protein | | Authors: | Ramelot, T.A, Cort, J.R, Zhao, L, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-23 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of HTH_XRE family transcriptional regulator BT_p548217 from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR244.
To be Published
|
|
2KOB
 
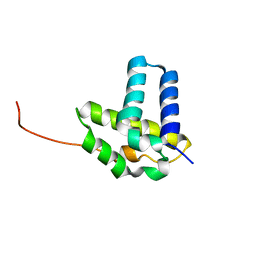 | | Solution NMR structure of CLOLEP_01837 (fragment 61-160) from Clostridium leptum. Northeast Structural Genomics Consortium Target QlR8A | | Descriptor: | Uncharacterized protein | | Authors: | Ramelot, T.A, Lee, D, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of CLOLEP_01837 (fragment 61-160) from Clostridium leptum. Northeast Structural Genomics Consortium Target QlR8A
To be Published
|
|
2KRS
 
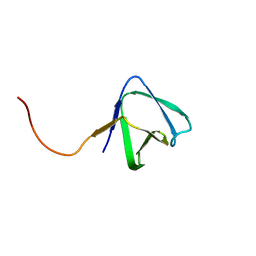 | | Solution NMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A. | | Descriptor: | Probable enterotoxin | | Authors: | Ramelot, T.A, Cort, J.R, Maglaqui, M, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A.
To be Published
|
|
2LGH
 
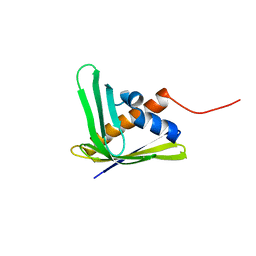 | | Solution NMR structure of the AHSA1-like protein AHA_2358 from Aeromonas hydrophila refined with NH RDCs, Northeast Structural Genomics Consortium Target AhR99. | | Descriptor: | Uncharacterized protein | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Wang, D, Ciccosanti, C, Janjua, H, Nair, R, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-26 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the AHSA1-like protein AHA_2358 from Aeromonas
hydrophila refined with NH RDCs. Northeast Structural Genomics Consortium Target AhR99.
To be Published
|
|
1R9P
 
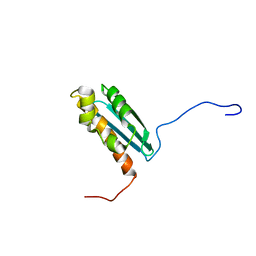 | | Solution NMR Structure Of The Haemophilus Influenzae Iron-Sulfur Cluster Assembly Protein U (IscU) with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target IR24. | | Descriptor: | NifU-like protein, ZINC ION | | Authors: | Ramelot, T.A, Cort, J.R, Xiao, R, Shastry, R, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-10-30 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site.
J.Mol.Biol., 344, 2004
|
|
1SGO
 
 | | NMR Structure of the human C14orf129 gene product, HSPC210. Northeast Structural Genomics target HR969. | | Descriptor: | Protein C14orf129 | | Authors: | Ramelot, T.A, Cort, J.R, Xiao, R, Shih, L.-Y, Ma, L.-C, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the human C14orf129 gene product, HSPC210. Northeast Structural Genomics target HR969.
To be Published
|
|
2KPI
 
 | | Solution NMR structure of Streptomyces coelicolor SCO3027 modeled with Zn+2 bound, Northeast Structural Genomics Consortium Target RR58 | | Descriptor: | Uncharacterized protein SCO3027, ZINC ION | | Authors: | Ramelot, T.A, Cort, J.R, Garcia, M, Yee, A, Arrowmith, C.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Streptomyces coelicolor SCO3027 modeled with Zn+2 bound, Northeast Structural Genomics Consortium Target RR58
To be Published
|
|
2LNA
 
 | | Solution NMR Structure of the mitochondrial inner membrane domain (residues 164-251), FtsH_ext, from the paraplegin-like protein AFG3L2 from Homo sapiens, Northeast Structural Genomics Consortium Target HR6741A | | Descriptor: | AFG3-like protein 2 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Janua, H, Kohan, E, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and MD simulations of the AAA protease intermembrane space domain indicates peripheral membrane localization within the hexaoligomer.
Febs Lett., 587, 2013
|
|
2B3W
 
 | | NMR structure of the E.coli protein YbiA, Northeast Structural Genomics target ET24. | | Descriptor: | Hypothetical protein ybiA | | Authors: | Ramelot, T.A, Cort, J.R, Xiao, R, Shih, L.Y, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-21 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the E.coli protein YbiA, Northeast Structural Genomics target ET24.
To be Published
|
|
2DSM
 
 | | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450 | | Descriptor: | Hypothetical protein yqaI | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-01 | | Release date: | 2006-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450
to be published
|
|
1YWU
 
 | | Solution NMR structure of Pseudomonas Aeruginosa protein PA4608. Northeast Structural Genomics target PaT7 | | Descriptor: | hypothetical protein PA4608 | | Authors: | Ramelot, T.A, Yee, A.A, Cort, J.R, Semesi, A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein.
Proteins, 66, 2007
|
|
7UBF
 
 | |
7UBD
 
 | |
