1T7I
 
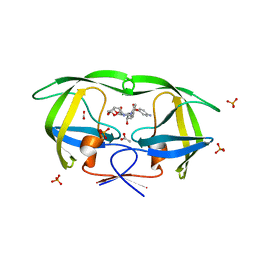 | | The structural and thermodynamic basis for the binding of TMC114, a next-generation HIV-1 protease inhibitor. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | King, N.M, Prabu-Jeyabalan, M, Nalivaika, E.A, Wigerinck, P.B.T.P, De Bethune, M.-P, Schiffer, C.A. | | Deposit date: | 2004-05-10 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor
J.Med.Chem., 48, 2005
|
|
3D3T
 
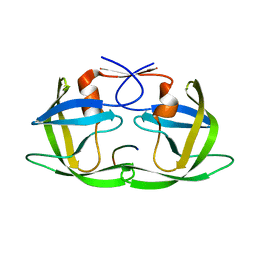 | | Crystal Structure of HIV-1 CRF01_AE in complex with the substrate p1-p6 | | Descriptor: | HIV-1 protease, P1-P6 SUBSTRATE PEPTIDE | | Authors: | Bandaranayake, R.M, Prabu-Jeyabalan, M, Kakizawa, J, Sugiura, W, Schiffer, C.A. | | Deposit date: | 2008-05-12 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of human immunodeficiency virus type 1 CRF01_AE protease in complex with the substrate p1-p6.
J.Virol., 82, 2008
|
|
3EL5
 
 | |
1F7A
 
 | |
2FNT
 
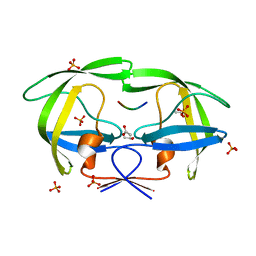 | | Crystal structure of a drug-resistant (V82A) inactive (D25N) HIV-1 protease complexed with AP2V variant of HIV-1 NC-p1 substrate. | | Descriptor: | ACETATE ION, NC-p1 substrate PEPTIDE, PHOSPHATE ION, ... | | Authors: | Prabu-Jeyabaln, M, Nalivaika, E.A, Schiffer, C.A. | | Deposit date: | 2006-01-11 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Mechanism of substrate recognition by drug-resistant human immunodeficiency virus type 1 protease variants revealed by a novel structural intermediate.
J.Virol., 80, 2006
|
|
1KJ7
 
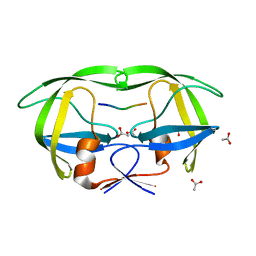 | |
1KJF
 
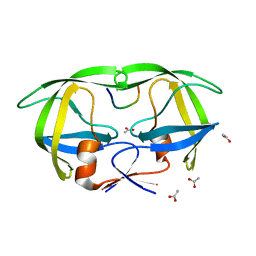 | |
1KJG
 
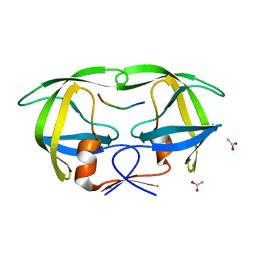 | |
1KJ4
 
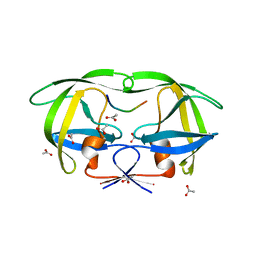 | |
1KJH
 
 | |
3OXX
 
 | | Crystal Structure of HIV-1 I50V, A71V Protease in Complex with the Protease Inhibitor Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, 1,2-ETHANEDIOL, ... | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-09-22 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50.
J.Virol., 87, 2013
|
|
3OXV
 
 | | Crystal Structure of HIV-1 I50V, A71 Protease in Complex with the protease inhibitor amprenavir. | | Descriptor: | ACETATE ION, GLYCEROL, HIV-1 Protease, ... | | Authors: | Schiffer, C.A, Mittal, S, Bandaranayake, R.M. | | Deposit date: | 2010-09-22 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50.
J.Virol., 87, 2013
|
|
3OXW
 
 | | Crystal Structure of HIV-1 I50V, A71V Protease in Complex with the Protease Inhibitor Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Mittal, S, Bandaranayake, R.M, Schiffer, C.A. | | Deposit date: | 2010-09-22 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50.
J.Virol., 87, 2013
|
|
1K6T
 
 | | LACK OF SYNERGY FOR INHIBITORS TARGETING A MULTI-DRUG RESISTANT HIV-1 PROTEASE | | Descriptor: | ACETATE ION, N-[2-HYDROXY-1-INDANYL]-5-[(2-TERTIARYBUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4-HYDROXY-2-(1-PHENYLETHYL)-PEN TANAMIDE, POL polyprotein | | Authors: | Schiffer, C.A. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Lack of synergy for inhibitors targeting a multi-drug-resistant HIV-1 protease.
Protein Sci., 11, 2002
|
|
1K6C
 
 | | LACK OF SYNERGY FOR INHIBITORS TARGETING A MULTI-DRUG RESISTANT HIV-1 PROTEASE | | Descriptor: | ACETATE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, POL polyprotein | | Authors: | Schiffer, C.A. | | Deposit date: | 2001-10-15 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lack of synergy for inhibitors targeting a multi-drug-resistant HIV-1 protease.
Protein Sci., 11, 2002
|
|
1K6P
 
 | | LACK OF SYNERGY FOR INHIBITORS TARGETING A MULTI-DRUG RESISTANT HIV-1 PROTEASE | | Descriptor: | ACETATE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(BENZO[1,3]DIOXOL-5-YLMETHYL)-PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, POL polyprotein | | Authors: | Schiffer, C.A. | | Deposit date: | 2001-10-16 | | Release date: | 2002-02-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lack of synergy for inhibitors targeting a multi-drug-resistant HIV-1 protease.
Protein Sci., 11, 2002
|
|
1K6V
 
 | | LACK OF SYNERGY FOR INHIBITORS TARGETING A MULTI-DRUG RESISTANT HIV-1 PROTEASE | | Descriptor: | ACETATE ION, N-[2-HYDROXY-1-INDANYL]-5-[(2-TERTIARYBUTYLAMINOCARBONYL)-4(BENZO[1,3]DIOXOL-5-YLMETHYL)-PIPERAZINO]-4-HYDROXY-2-(1-PHE NYLETHYL)-PENTANAMIDE, POL polyprotein | | Authors: | Schiffer, C.A. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lack of synergy for inhibitors targeting a multi-drug-resistant HIV-1 protease.
Protein Sci., 11, 2002
|
|
3EKP
 
 | | Crystal Structure of the inhibitor Amprenavir (APV) in complex with a multi-drug resistant HIV-1 protease variant (L10I/G48V/I54V/V64I/V82A)Refer: FLAP+ in citation | | Descriptor: | ACETATE ION, PHOSPHATE ION, Protease, ... | | Authors: | Prabu-Jayabalan, M, King, N.M, Bandaranayake, R.M. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
3EKX
 
 | |
3EKV
 
 | |
3EL1
 
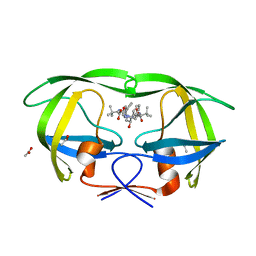 | | Crystal Structure of wild-type HIV protease in complex with the inhibitor, Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
3EKY
 
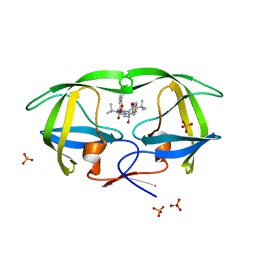 | | Crystal Structure of wild-type HIV protease in complex with the inhibitor, Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
