4ISJ
 
 | | RNA Ligase RtcB in complex with Mn(II) | | Descriptor: | MANGANESE (II) ION, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.344 Å) | | Cite: | Structures of the Noncanonical RNA Ligase RtcB Reveal the Mechanism of Histidine Guanylylation.
Biochemistry, 52, 2013
|
|
4IT0
 
 | | Structure of the RNA ligase RtcB-GMP/Mn(II) complex | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the Noncanonical RNA Ligase RtcB Reveal the Mechanism of Histidine Guanylylation.
Biochemistry, 52, 2013
|
|
4ISZ
 
 | | RNA ligase RtcB in complex with GTP alphaS and Mn(II) | | Descriptor: | GUANOSINE-5'-RP-ALPHA-THIO-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-01-17 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structures of the Noncanonical RNA Ligase RtcB Reveal the Mechanism of Histidine Guanylylation.
Biochemistry, 52, 2013
|
|
4K0B
 
 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-01 | | Last modified: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4L7I
 
 | | Crystal structure of S-Adenosylmethionine synthase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4L2Z
 
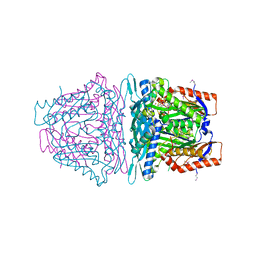 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAE and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-05 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4ZAH
 
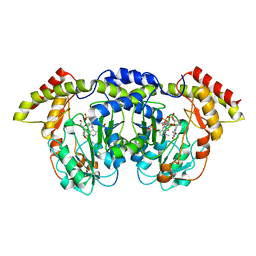 | | Crystal structure of sugar aminotransferase WecE with External Aldimine VII from Escherichia coli K-12 | | Descriptor: | [[(2R,3S,5R)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3R,4S,5R,6R)-6-methyl-5-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3,4-bis(oxidanyl)oxan-2-yl] hydrogen phosphate, dTDP-4-amino-4,6-dideoxygalactose transaminase | | Authors: | Wang, F, Singh, S, Cao, H, Xu, W, Miller, M.D, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Basis for the Stereochemical Control of Amine Installation in Nucleotide Sugar Aminotransferases.
Acs Chem.Biol., 10, 2015
|
|
4ZDN
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-17 | | Release date: | 2015-05-13 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | Descriptor: | GLYCEROL, Putative aminotransferase | | Authors: | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-19 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
5D6Y
 
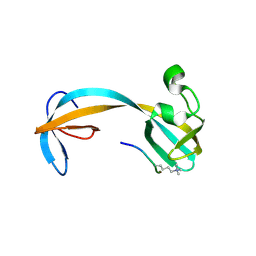 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A complexed with histone H3K23me3 | | Descriptor: | Lysine-specific demethylase 4A, peptide H3K23me3 (19-28) | | Authors: | Wang, F, Su, Z, Miller, M.D, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2016-02-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
5D6W
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A | | Descriptor: | Lysine-specific demethylase 4A, S,R MESO-TARTARIC ACID | | Authors: | Wang, F, Su, Z, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
5D6X
 
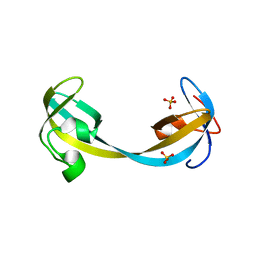 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A | | Descriptor: | Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Wang, F, Su, Z, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
5E7Q
 
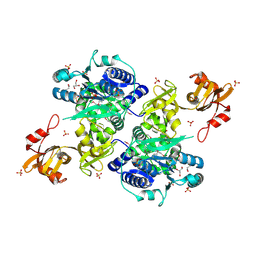 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis | | Descriptor: | GLYCEROL, SULFATE ION, acyl-CoA synthetase | | Authors: | Osipiuk, J, Cuff, M.E, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J, Ma, M, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-12 | | Release date: | 2015-10-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
4MV2
 
 | | Crystal structure of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 | | Authors: | Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Thomas, M.G, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-02 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
4NI3
 
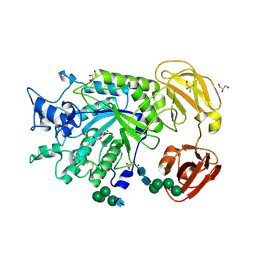 | | Crystal Structure of GH29 family alpha-L-fucosidase from Fusarium graminearum in the closed form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-fucosidase GH29, ... | | Authors: | Cao, H, Walton, J.D, Brumm, P, Phillips Jr, G.N. | | Deposit date: | 2013-11-05 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3993 Å) | | Cite: | Structure and Substrate Specificity of a Eukaryotic Fucosidase from Fusarium graminearum.
J.Biol.Chem., 289, 2014
|
|
4NEO
 
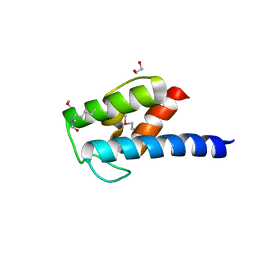 | | Structure of BlmI, a type-II acyl-carrier-protein from Streptomyces verticillus involved in bleomycin biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Peptide synthetase NRPS type II-PCP | | Authors: | Cuff, M.E, Bigelow, L, Bearden, J, Babnigg, G, Bruno, C.J.P, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of BlmI as a model for nonribosomal peptide synthetase peptidyl carrier proteins.
Proteins, 82, 2014
|
|
4OPF
 
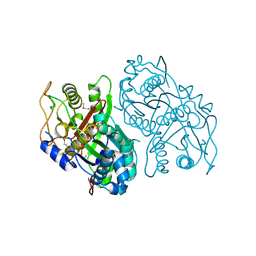 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS8 | | Descriptor: | NRPS/PKS | | Authors: | Osipiuk, J, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8EBR
 
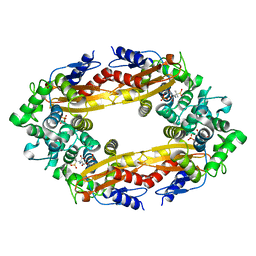 | |
8EC4
 
 | |
8EBI
 
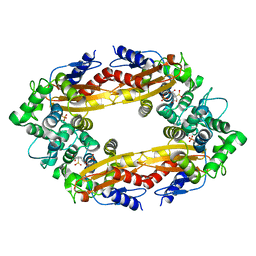 | |
8ECF
 
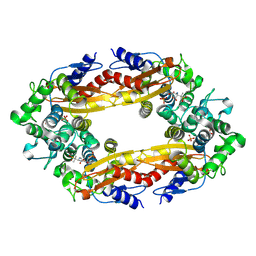 | |
4OO2
 
 | | Streptomyces globisporus C-1027 FAD dependent (S)-3-chloro-β-tyrosine-S-SgcC2 C-5 hydroxylase SgcC apo form | | Descriptor: | CALCIUM ION, Chlorophenol-4-monooxygenase, GLYCEROL | | Authors: | Cao, H, Xu, W, Bingman, C.A, Lohman, J.R, Yennamalli, R, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-01-29 | | Release date: | 2014-02-12 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
4OPE
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS7 | | Descriptor: | NITRATE ION, NRPS/PKS | | Authors: | Osipiuk, J, Mack, J, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4OQJ
 
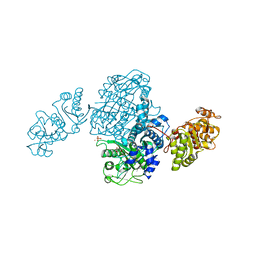 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmQ KS1 | | Descriptor: | GLYCEROL, PHOSPHATE ION, PKS, ... | | Authors: | Nocek, B, Mack, J, Endras, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-09 | | Release date: | 2014-03-19 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4OVM
 
 | | Crystal structure of SgcJ protein from Streptomyces carzinostaticus | | Descriptor: | uncharacterized protein SgcJ | | Authors: | Chang, C, Bigelow, L, Clancy, S, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-25 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.719 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J.Antibiot., 2016
|
|
