2OVQ
 
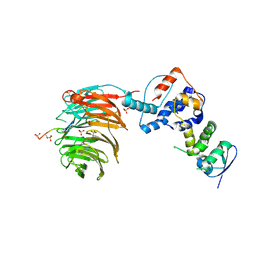 | | Structure of the Skp1-Fbw7-CyclinEdegC complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A, SULFATE ION, ... | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
2OVP
 
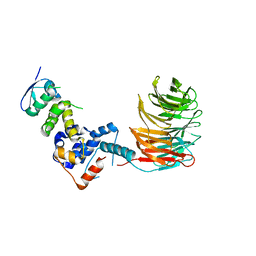 | | Structure of the Skp1-Fbw7 complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
2QSH
 
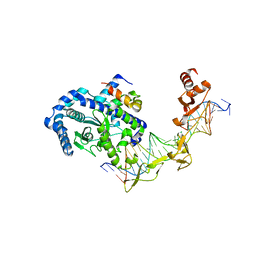 | | Crystal structure of Rad4-Rad23 bound to a mismatch DNA | | Descriptor: | DNA repair protein RAD4, UV excision repair protein RAD23, bottom strand of the mismatch DNA, ... | | Authors: | Min, J.-H, Pavletich, N.P. | | Deposit date: | 2007-07-31 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Recognition of DNA damage by the Rad4 nucleotide excision repair protein
Nature, 449, 2007
|
|
2QSF
 
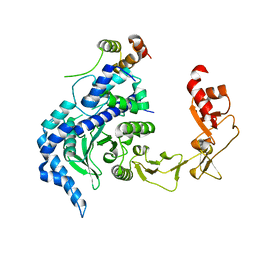 | | Crystal structure of the Rad4-Rad23 complex | | Descriptor: | DNA repair protein RAD4, UV excision repair protein RAD23 | | Authors: | Min, J.-H, Pavletich, N.P. | | Deposit date: | 2007-07-31 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Recognition of DNA damage by the Rad4 nucleotide excision repair protein
Nature, 449, 2007
|
|
2QSG
 
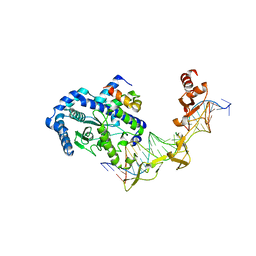 | |
5EAY
 
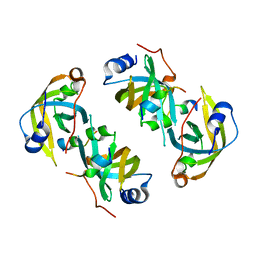 | |
5EAN
 
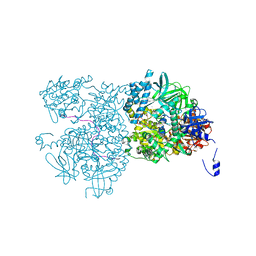 | | Crystal structure of Dna2 in complex with a 5' overhang DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*AP*CP*TP*CP*TP*GP*CP*CP*AP*AP*GP*AP*GP*GP*A)-3'), ... | | Authors: | Zhou, C, Pourmal, S, Pavletich, N.P. | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
5EAX
 
 | | Crystal structure of Dna2 in complex with an ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication ATP-dependent helicase/nuclease DNA2, ... | | Authors: | Zhou, C, Pourmal, S, Pavletich, N.P. | | Deposit date: | 2015-10-17 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
5EAW
 
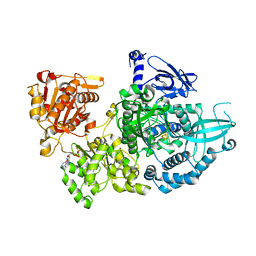 | | Crystal structure of Dna2 nuclease-helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication ATP-dependent helicase/nuclease DNA2, IRON/SULFUR CLUSTER | | Authors: | Zhou, C, Pourmal, S, Pavletich, N.P. | | Deposit date: | 2015-10-17 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
1YER
 
 | |
1YET
 
 | |
1YCR
 
 | |
1Z3I
 
 | | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | | Descriptor: | SULFATE ION, ZINC ION, similar to RAD54-like | | Authors: | Thoma, N.H, Czyzewski, B.K, Alexeev, A.A, Mazin, A.V, Kowalczykowski, S.C, Pavletich, N.P. | | Deposit date: | 2005-03-12 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YGS
 
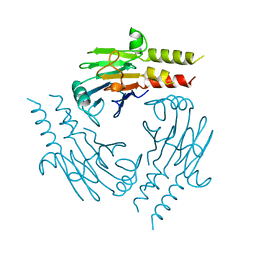 | | CRYSTAL STRUCTURE OF THE SMAD4 TUMOR SUPPRESSOR C-TERMINAL DOMAIN | | Descriptor: | SMAD4 | | Authors: | Shi, Y, Hata, A, Lo, R.S, Massague, J, Pavletich, N.P. | | Deposit date: | 1997-10-03 | | Release date: | 1998-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for mutational inactivation of the tumour suppressor Smad4.
Nature, 388, 1997
|
|
1YCQ
 
 | |
1YES
 
 | |
1YCS
 
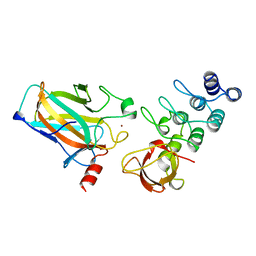 | | P53-53BP2 COMPLEX | | Descriptor: | 53BP2, P53, ZINC ION | | Authors: | Gorina, S, Pavletich, N.P. | | Deposit date: | 1996-09-30 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2.
Science, 274, 1996
|
|
1T4W
 
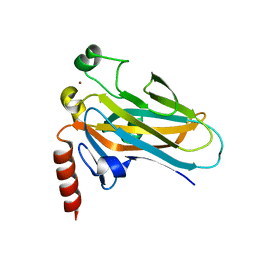 | | Structural Differences in the DNA Binding Domains of Human p53 and its C. elegans Ortholog Cep-1: Structure of C. elegans Cep-1 | | Descriptor: | C.Elegans p53 tumor suppressor-like transcription factor, ZINC ION | | Authors: | Huyen, Y, Jeffrey, P.D, Derry, W.B, Rothman, J.H, Pavletich, N.P, Stavridi, E.S, Halazonetis, T.D. | | Deposit date: | 2004-04-30 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Differences in the DNA Binding Domains of Human p53 and Its C. elegans Ortholog Cep-1.
Structure, 12, 2004
|
|
1VCB
 
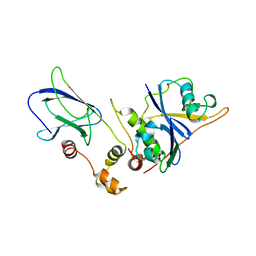 | | THE VHL-ELONGINC-ELONGINB STRUCTURE | | Descriptor: | PROTEIN (ELONGIN B), PROTEIN (ELONGIN C), PROTEIN (VHL) | | Authors: | Stebbins, C.E, Kaelin, W.G, Pavletich, N.P. | | Deposit date: | 1999-03-13 | | Release date: | 1999-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function.
Science, 284, 1999
|
|
2ASS
 
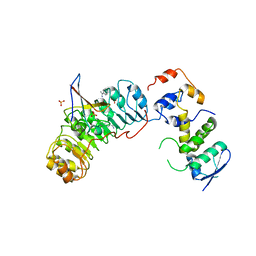 | | Crystal structure of the Skp1-Skp2-Cks1 complex | | Descriptor: | BENZAMIDINE, Cyclin-dependent kinases regulatory subunit 1, PHOSPHATE ION, ... | | Authors: | Hao, B, Zhang, N, Schulman, B.A, Wu, G, Pagano, M, Pavletich, N.P. | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of the Cks1-Dependent Recognition of p27(Kip1) by the SCF(Skp2) Ubiquitin Ligase.
Mol.Cell, 20, 2005
|
|
2AZE
 
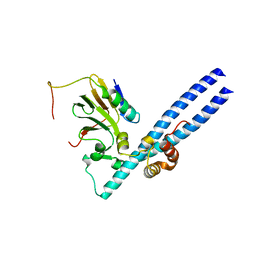 | | Structure of the Rb C-terminal domain bound to an E2F1-DP1 heterodimer | | Descriptor: | Retinoblastoma-associated protein, Transcription factor Dp-1, Transcription factor E2F1 | | Authors: | Rubin, S.M, Gall, A.L, Zheng, N, Pavletich, N.P. | | Deposit date: | 2005-09-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release.
Cell(Cambridge,Mass.), 123, 2005
|
|
1OZJ
 
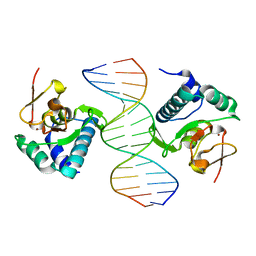 | | Crystal structure of Smad3-MH1 bound to DNA at 2.4 A resolution | | Descriptor: | SMAD 3, Smad binding element, ZINC ION | | Authors: | Chai, J, Wu, J.-W, Yan, N, Massague, J, Pavletich, N.P, Shi, Y. | | Deposit date: | 2003-04-09 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Features of a Smad3 MH1-DNA complex. Roles of water and zinc in DNA binding.
J.Biol.Chem., 278, 2003
|
|
1MP8
 
 | | Crystal structure of Focal Adhesion Kinase (FAK) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, focal adhesion kinase 1 | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C.G, Pavletich, N.P, Rodgers, J, Sang, B.-C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-11 | | Release date: | 2003-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography
Structure, 10, 2002
|
|
1P22
 
 | | Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity on the SCFbeta-TrCP1 ubiquitin ligase | | Descriptor: | Beta-catenin, F-box/WD-repeat protein 1A, Skp1 | | Authors: | Wu, G, Xu, G, Schulman, B.A, Jeffrey, P.D, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a beta-TrCP1-Skp1-beta-Catenin complex: destruction motif binding and lysine specificity of the SCFbeta-TrCP1 ubiquitin ligase
Mol.Cell, 11, 2003
|
|
1MJE
 
 | | STRUCTURE OF A BRCA2-DSS1-SSDNA COMPLEX | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*T)-3', Deleted in split hand/split foot protein 1, breast cancer 2 | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-27 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure.
Science, 297, 2002
|
|
