2VR5
 
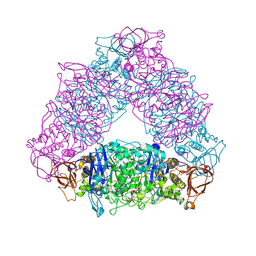 | | Crystal structure of Trex from Sulfolobus Solfataricus in complex with acarbose intermediate and glucose | | Descriptor: | 4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranosyl)-beta-D-glucopyranose, GLYCEROL, GLYCOGEN OPERON PROTEIN GLGX, ... | | Authors: | Song, H.-N, Yoon, S.-M, Lee, S.-J, Cha, H.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2008-03-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
2VUY
 
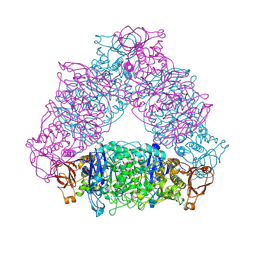 | | Crystal structure of Glycogen Debranching exzyme TreX from Sulfolobus solfatarius | | Descriptor: | GLYCOGEN OPERON PROTEIN GLGX | | Authors: | Song, H.-N, Yoon, S.-M, Cha, H.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
7WKR
 
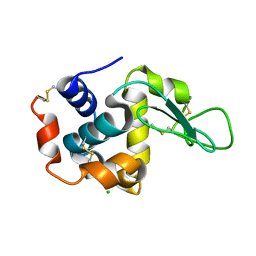 | |
7WUC
 
 | |
3ZHL
 
 | |
4RF3
 
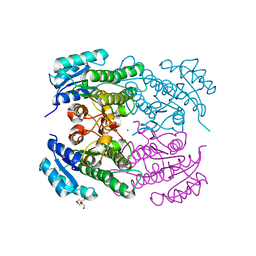 | | Crystal Structure of ketoreductase from Lactobacillus kefir, mutant A94F | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADPH dependent R-specific alcohol dehydrogenase | | Authors: | Tang, Y, Tibrewal, N, Cascio, D. | | Deposit date: | 2014-09-24 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Origins of stereoselectivity in evolved ketoreductases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RF5
 
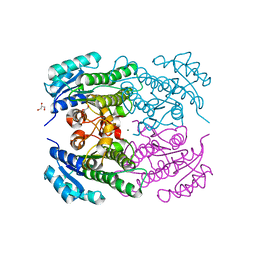 | | Crystal structure of ketoreductase from Lactobacillus kefir, E145S mutant | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADPH dependent R-specific alcohol dehydrogenase | | Authors: | Tang, Y, Tibrewal, N, Cascio, D. | | Deposit date: | 2014-09-24 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Origins of stereoselectivity in evolved ketoreductases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RF2
 
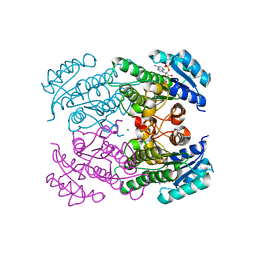 | |
4RF4
 
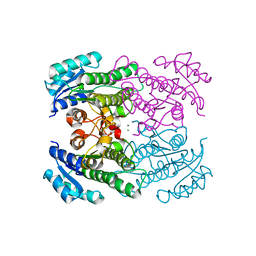 | |
6XCS
 
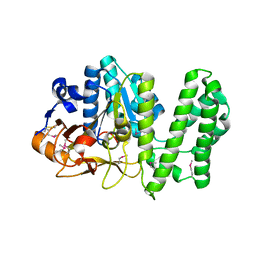 | |
4TN5
 
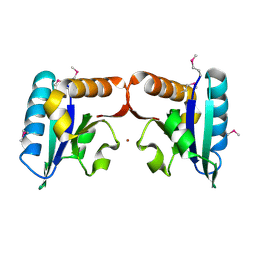 | |
8SRP
 
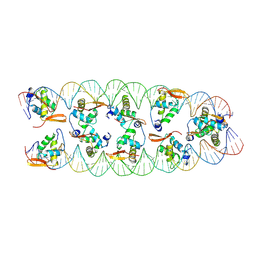 | |
8SRO
 
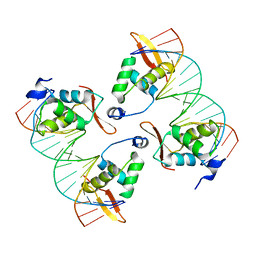 | | FoxP3 tetramer on TTTG repeats | | Descriptor: | DNA 72-mer, Forkhead box protein P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | FOXP3 recognizes microsatellites and bridges DNA through multimerization.
Nature, 624, 2023
|
|
6JCG
 
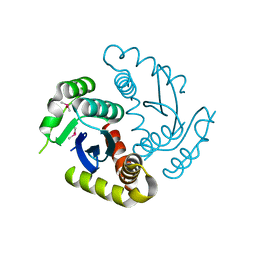 | | Room temperature structure of HIV-1 Integrase catalytic core domain by serial femtosecond crystallography. | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Shi, Y, Han, J, Li, X, Kim, T.H, Yun, J.H. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
6JCF
 
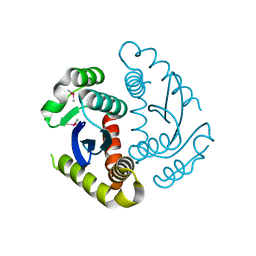 | | Cryogenic structure of HIV-1 Integrase catalytic core domain by synchrotron | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Han, J, Kim, T.H, Yun, J.H, Lee, W. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
7X9R
 
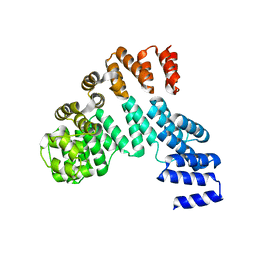 | | Crystal structure of the antirepressor GmaR | | Descriptor: | Glycosyl transferase family 2 | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
7X9S
 
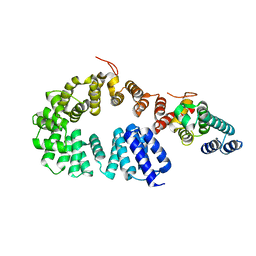 | | Crystal structure of a complex between the antirepressor GmaR and the transcriptional repressor MogR | | Descriptor: | GmaR, Motility gene repressor MogR | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
8GUW
 
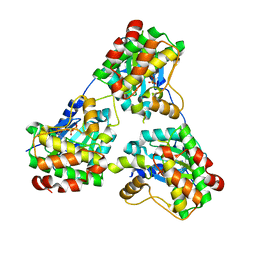 | |
6VR3
 
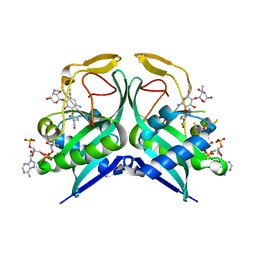 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with acetylated-netilmicin and CoA | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, N-[(2S,3R)-2-{[(1R,2S,3S,4R,6S)-6-amino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-lyxopyranosyl]oxy}-4-(ethylamino) -2-hydroxycyclohexyl]oxy}-6-(aminomethyl)-3,4-dihydro-2H-pyran-3-yl]acetamide | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-02-06 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
6VR2
 
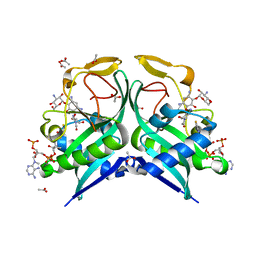 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with acetylated-tobramycin and CoA | | Descriptor: | (1S,2S,3R,4S,6R)-3-{[2-(acetylamino)-6-amino-2,3,6-trideoxy-alpha-D-ribo-hexopyranosyl]oxy}-4,6-diamino-2-hydroxycycloh exyl 3-amino-3-deoxy-alpha-D-glucopyranoside, ACETATE ION, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-02-06 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
6VTA
 
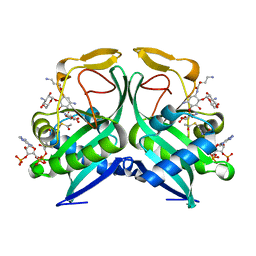 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with amikacin and acetyl-CoA | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, ACETYL COENZYME *A, Aminoglycoside 2'-N-acetyltransferase | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-02-12 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
7YCJ
 
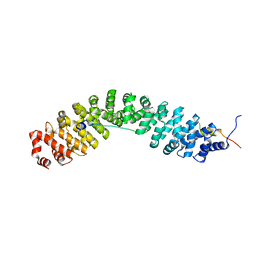 | |
4E0N
 
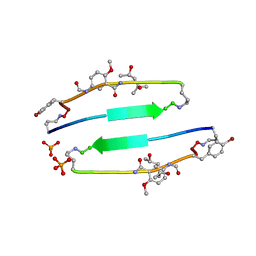 | | SVQIVYK segment from human Tau (305-311) displayed on 54-membered macrocycle scaffold (form II) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide SVQIVYK(ORN)EF(HAO)(4BF)K(ORN), PHOSPHATE ION | | Authors: | Zhao, M, Liu, C, Michael, S.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4E0O
 
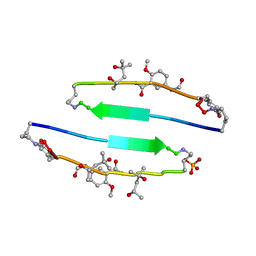 | | SVQIVYK segment from human Tau (305-311) displayed on 54-membered macrocycle scaffold (form III) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide SVQIVYK(ORN)EF(HAO)(4BF)K(ORN), PHOSPHATE ION | | Authors: | Zhao, M, Liu, C, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8K6U
 
 | |
