4GVJ
 
 | | Tyk2 (JH1) in complex with adenosine di-phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Liang, J, Abbema, A.V, Bao, L, Barrett, K, Beresini, M, Berezhkovskiy, L, Blair, W, Chang, C, Driscoll, J, Eigenbrot, C, Ghilardi, N, Gibbons, P, Halladay, J, Johnson, A, Kohli, P.B, Lai, Y, Liimatta, M, Mantik, P, Menghrajani, K, Murray, J, Sambrone, A, Shao, Y, Shia, S, Shin, Y, Smith, J, Sohn, S, Stanley, M, Tsui, V, Ultsch, M, Wu, L, Zhang, B, Magnuson, S. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
4FCP
 
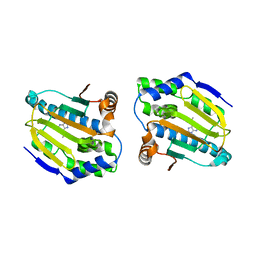 | | Targetting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo [2,3-d] pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization | | Descriptor: | Heat shock protein HSP 90-alpha, N,N-dimethyl-7H-purin-6-amine | | Authors: | Davies, N.G.M, Browne, H, Davies, B, Foloppe, N, Geoffrey, S, Gibbons, B, Hart, T, Drysdale, M, Mansell, H, Massey, A, Matassova, N, Moore, J.D, Murray, J, Pratt, R, Ray, S, Roughley, S.D, Jensen, M.R, Schoepfer, J, Scriven, K, Simmonite, H, Stokes, S, Surgenor, A, Webb, P, Wright, L, Brough, P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization.
Bioorg.Med.Chem., 20, 2012
|
|
4FCR
 
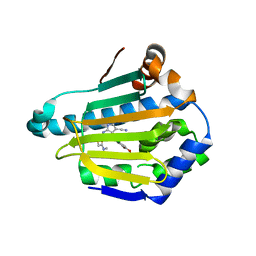 | | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization | | Descriptor: | 2-{[4-(2-chloro-4,5-dimethoxyphenyl)-5-cyano-7H-pyrrolo[2,3-d]pyrimidin-2-yl]sulfanyl}-N,N-dimethylacetamide, Heat shock protein HSP 90-alpha | | Authors: | Davies, N.G, Browne, H, Davis, B, Foloppe, N, Geoffrey, S, Gibbons, B, Hart, T, Drysdale, M.J, Mansell, H, Massey, A, Matassova, N, Moore, J.D, Murray, J, Pratt, R, Ray, S, Roughley, S.D, Jensen, M.R, Schoepfer, J, Scriven, K, Simmonite, H, Stokes, S, Surgenor, A, Webb, P, Wright, L, Brough, P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization.
Bioorg.Med.Chem., 20, 2012
|
|
5LOF
 
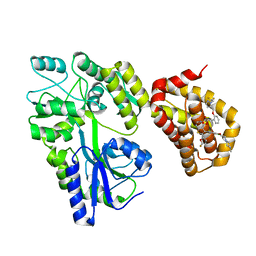 | | Crystal structure of the MBP-MCL1 complex with highly selective and potent inhibitor of MCL1 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(5-fluoranylfuran-2-yl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-[2,2,2-tris(fluoranyl)ethyl]pyrazol-3-yl]methoxy]phenyl]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Kotschy, A, Szlavik, Z, Murray, J, Davidson, J, Csekei, M, Paczal, A, Szabo, Z, Sipos, S, Radics, G, Proszenyak, A, Balint, B, Ondi, L, Blasko, G, Robertson, A, Surgenor, A, Chen, I, Matassova, N, Smith, J, Pedder, C, Graham, C, Geneste, O. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models.
Nature, 538, 2016
|
|
4FCQ
 
 | | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization | | Descriptor: | 4-(2,4-dimethylphenyl)-2-(methylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha | | Authors: | Davies, N.G, Browne, H, Davis, B, Foloppe, N, Geoffrey, S, Gibbons, B, Hart, T, Drysdale, M.J, Mansell, H, Massey, A, Matassova, N, Moore, J.D, Murray, J, Pratt, R, Ray, S, Roughley, S.D, Jensen, M.R, Schoepfer, J, Scriven, K, Simmonite, H, Stokes, S, Surgenor, A, Webb, P, Wright, L, Brough, P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization.
Bioorg.Med.Chem., 20, 2012
|
|
5EMW
 
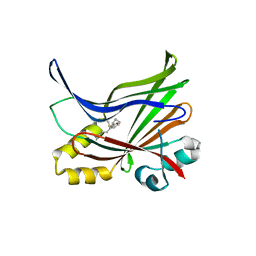 | | Crystal structure of the palmitoylated human TEAD3 transcription factor | | Descriptor: | CALCIUM ION, Transcriptional enhancer factor TEF-5 | | Authors: | Noland, C.L, Gierke, S, Schnier, P.D, Murray, J, Sandoval, W.N, Sagolla, M, Dey, A, Hannoush, R.N, Fairbrother, W.J, Cunningham, C.N. | | Deposit date: | 2015-11-06 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Palmitoylation of TEAD Transcription Factors Is Required for Their Stability and Function in Hippo Pathway Signaling.
Structure, 24, 2016
|
|
5FND
 
 | | Dynamic Undocking and the Quasi-Bound State as tools for Drug Design | | Descriptor: | HEAT SHOCK PROTEIN, HSP90-ALPHA, N-(2-AZANYL-6-METHYL-1,3-BENZOTHIAZOL-5-YL)ETHANAMIDE, ... | | Authors: | Ruiz-Carmona, S, Schmidtke, P, Luque, F.J, Baker, L.M, Matassova, N, Davis, B, Roughley, S, Murray, J, Hubbard, R, Barril, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dynamic undocking and the quasi-bound state as tools for drug discovery.
Nat Chem, 9, 2017
|
|
5FNF
 
 | | Dynamic Undocking and the Quasi-Bound State as tools for Drug Design | | Descriptor: | 4-[(E)-N-oxidanyl-C-pyridin-3-yl-carbonimidoyl]benzene-1,3-diol, HEAT SHOCK PROTEIN, HSP90-ALPHA, ... | | Authors: | Ruiz-Carmona, S, Schmidtke, P, Luque, F.J, Baker, L.M, Matassova, N, Davis, B, Roughley, S, Murray, J, Hubbard, R, Barril, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dynamic undocking and the quasi-bound state as tools for drug discovery.
Nat Chem, 9, 2017
|
|
5FNC
 
 | | Dynamic Undocking and the Quasi-Bound State as tools for Drug Design | | Descriptor: | 6-CHLORO-4-N-[(4-METHYLPHENYL)METHYL]PYRIMIDINE- 2,4-DIAMINE, HEAT SHOCK PROTEIN, HSP90-ALPHA, ... | | Authors: | Ruiz-Carmona, S, Schmidtke, P, Luque, F.J, Baker, L.M, Matassova, N, Davis, B, Roughley, S, Murray, J, Hubbard, R, Barril, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic undocking and the quasi-bound state as tools for drug discovery.
Nat Chem, 9, 2017
|
|
5EMV
 
 | | Crystal structure of the palmitoylated human TEAD2 transcription factor | | Descriptor: | Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Gierke, S, Schnier, P.D, Murray, J, Sandoval, W.N, Sagolla, M, Dey, A, Hannoush, R.N, Fairbrother, W.J, Cunningham, C.N. | | Deposit date: | 2015-11-06 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Palmitoylation of TEAD Transcription Factors Is Required for Their Stability and Function in Hippo Pathway Signaling.
Structure, 24, 2016
|
|
3JAH
 
 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAG stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
3JAG
 
 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAA stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
3COS
 
 | | Crystal structure of human class II alcohol dehydrogenase (ADH4) in complex with NAD and Zn | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Alcohol dehydrogenase 4, ... | | Authors: | Kavanagh, K.L, Shafqat, N, Yue, W, von Delft, F, Bishop, S, Roos, A, Murray, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-29 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human class II alcohol dehydrogenase (ADH4) in complex with NAD and Zn.
To be Published
|
|
3JAI
 
 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UGA stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
6B8U
 
 | | Crystals Structure of B-Raf kinase domain in complex with an Imidazopyridinyl benzamide inhibitor | | Descriptor: | Serine/threonine-protein kinase B-raf, ~{N}-[3-(2-acetamidoimidazo[1,2-a]pyridin-6-yl)-4-methyl-phenyl]-3-(trifluoromethyl)benzamide | | Authors: | Appleton, B.A, Murray, J, Shafer, C.M. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Imidazo[1,2-a]pyridin-6-yl-benzamide analogs as potent RAF inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5LZV
 
 | | Structure of the mammalian ribosomal termination complex with accommodated eRF1(AAQ) and ABCE1. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5LZX
 
 | | Structure of the mammalian rescue complex with Pelota and Hbs1l assembled on a UGA stop codon. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5LZZ
 
 | | Structure of the mammalian rescue complex with Pelota and Hbs1l (combined) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5LZY
 
 | | Structure of the mammalian rescue complex with Pelota and Hbs1l assembled on a polyadenylated mRNA. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5LZW
 
 | | Structure of the mammalian rescue complex with Pelota and Hbs1l assembled on a truncated mRNA. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
2VRF
 
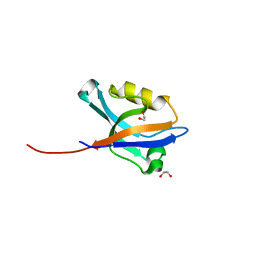 | | CRYSTAL STRUCTURE OF THE HUMAN BETA-2-SYNTROPHIN PDZ DOMAIN | | Descriptor: | 1,2-ETHANEDIOL, BETA-2-SYNTROPHIN | | Authors: | Sun, Z, Roos, A.K, Pike, A.C.W, Pilka, E.S, Cooper, C, Elkins, J.M, Murray, J, Arrowsmith, C.H, Doyle, D, Edwards, A, von Delft, F, Bountra, C, Oppermann, U. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Beta-2-Syntrophin Pdz Domain
To be Published
|
|
2VRE
 
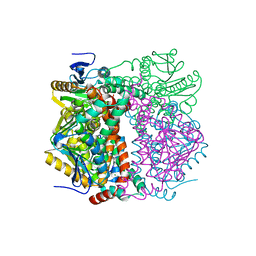 | | crystal structure of human peroxisomal delta3,5,delta2,4-dienoyl coa isomerase | | Descriptor: | CHLORIDE ION, DELTA(3,5)-DELTA(2,4)-DIENOYL-COA ISOMERASE | | Authors: | Yue, W, Guo, K, von Delft, F, Pilka, E, Murray, J, Roos, A, Kochan, G, Bountra, C, Arrowsmith, C, Wikstrom, M, Edwards, A, Oppermann, U. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Human Peroxisomal Delta3,5, Delta2,4-Dienoyl Coa Isomerase (Ech1)
To be Published
|
|
4NBK
 
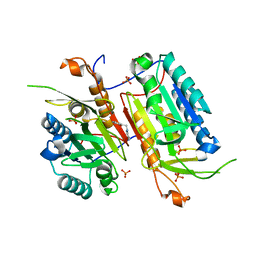 | |
4N7M
 
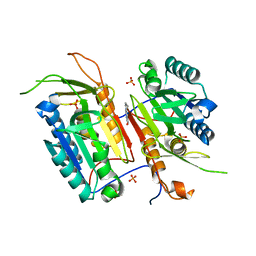 | |
4N6G
 
 | |
