5DF0
 
 | | Crystal structure of AcMNPV Chitinase A in complex WITH CHITOTRIO-THIAZOLINE DITHIOAMIDE | | Descriptor: | (2R,3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methylhexahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, ... | | Authors: | Mou, T.-C, Sprang, S.R. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Crystal structure of AcMNPV Chitinase A
To Be Published
|
|
2GVZ
 
 | |
2GVD
 
 | |
5DEZ
 
 | | Crystal structure of AcMNPV Chitinase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ac-ChiA, ... | | Authors: | Mou, T.-C, Sprang, S.R. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of AcMNPV Chitinase A
To Be Published
|
|
5I56
 
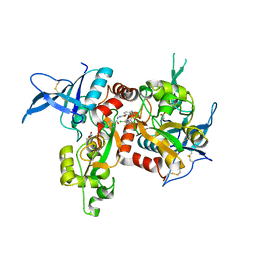 | | Agonist-bound GluN1/GluN2A agonist binding domains with TCN201 | | Descriptor: | GLUTAMIC ACID, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
3G82
 
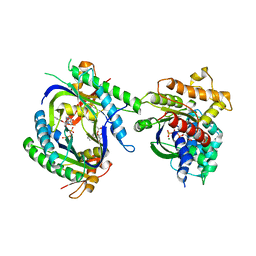 | | Complex of GS-alpha with the catalytic domains of mammalian adenylyl cyclase: complex with MANT-ITP and Mn | | Descriptor: | 3'-O-{[2-(methylamino)phenyl]carbonyl}inosine 5'-(tetrahydrogen triphosphate), 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase type 2, ... | | Authors: | Huebner, M, Mou, T.-C, Sprang, S.R, Seifert, R. | | Deposit date: | 2009-02-11 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | 2',3'-(O)-(N-Methyl)anthraniloyl-inosine 5'-triphosphate is the Most Potent Adenylyl Cyclase 1 and 5 Inhibitor Known so far and Effectively Promotes Catalytic Subunit Assembly in the Absence of Forskolin
To be Published
|
|
3MAA
 
 | |
5VIH
 
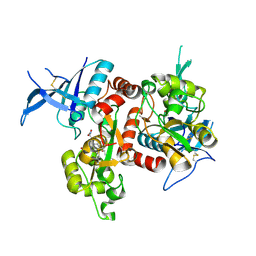 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-fluorophenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-(4-fluorophenyl)-1H-pyrazole-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCINE, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VIJ
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-bromophenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-(4-bromophenyl)-1H-pyrazole-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VII
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-(3-fluoropropyl)phenyl-ACEPC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(2R)-2-amino-2-carboxyethyl]-1-[4-(3-fluoropropyl)phenyl]-1H-pyrazole-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5DEX
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, phenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-phenyl-1H-pyrazole-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 2017
|
|
5I59
 
 | | Glutamate- and glycine-bound GluN1/GluN2A agonist binding domains with MPX 007 | | Descriptor: | 5-({[(3,4-difluorophenyl)sulfonyl]amino}methyl)-6-methyl-N-[(2-methyl-4H-1lambda~4~,3-thiazol-5-yl)methyl]pyrazine-2-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
5I57
 
 | | Glutamate- and glycine-bound GluN1/GluN2A agonist binding domains | | Descriptor: | GLUTAMIC ACID, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
5I58
 
 | | GLUTAMATE- AND GLYCINE-BOUND GLUN1/GLUN2A AGONIST BINDING DOMAINS WITH MPX-004 | | Descriptor: | 5-({[(3-chloro-4-fluorophenyl)sulfonyl]amino}methyl)-N-[(2-methyl-1,3-thiazol-5-yl)methyl]pyrazine-2-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
5JTY
 
 | | Glutamate- and DCKA-bound GluN1/GluN2A agonist binding domains with MPX-007 | | Descriptor: | 4-hydroxy-5,7-dimethylquinoline-2-carboxylic acid, 5-({[(3,4-difluorophenyl)sulfonyl]amino}methyl)-6-methyl-N-[(2-methyl-1,3-thiazol-5-yl)methyl]pyrazine-2-carboxamide, GLUTAMIC ACID, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-05-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
3C16
 
 | |
3C14
 
 | |
3C15
 
 | |
9BFB
 
 | | Crystal structure of BRAF kinase domain with PF-07284890 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-{2-chloro-3-[(3,5-dimethyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]-4-fluorophenyl}-3-fluoropropane-1-sulfonamide, ... | | Authors: | Mou, T.-C. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of the Clinical Candidate PF-07284890 ( ARRY-461 ), a Highly Potent and Brain Penetrant BRAF Inhibitor for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
4MU8
 
 | | Crystal structure of an oxidized form of yeast iso-1-cytochrome c at pH 8.8 | | Descriptor: | Cytochrome c iso-1, GLYCEROL, HEME C, ... | | Authors: | McClelland, L.J, Mou, T.-C, Jeakins-Cooley, M.E, Sprang, S.R, Bowler, B.E. | | Deposit date: | 2013-09-20 | | Release date: | 2014-06-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a mitochondrial cytochrome c conformer competent for peroxidase activity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
9C93
 
 | | Crystal structure of menin in complex with inhibitor compound 26 | | Descriptor: | (1R,5R)-2-({5-fluoro-2-[(5-{7-[(1-methylcyclopropyl)methyl]-2,7-diazaspiro[3.5]nonan-2-yl}-1,2,4-triazin-6-yl)oxy]phenyl}methyl)-2-azabicyclo[3.1.0]hexan-3-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Menin, ... | | Authors: | Bester, S.M, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of Potent Menin-KMT2A Interaction Inhibitors with Improved In Vitro ADME Properties and Reduced hERG Affinity.
Acs Med.Chem.Lett., 16, 2025
|
|
9C92
 
 | | Crystal structure of menin in complex with inhibitor compound 15 | | Descriptor: | Menin, N-ethyl-5-fluoro-2-[(5-{7-[(1-methylcyclopropyl)methyl]-2,7-diazaspiro[3.5]nonan-2-yl}-1,2,4-triazin-6-yl)oxy]-N-(propan-2-yl)benzamide | | Authors: | Bester, S.M, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Potent Menin-KMT2A Interaction Inhibitors with Improved In Vitro ADME Properties and Reduced hERG Affinity.
Acs Med.Chem.Lett., 16, 2025
|
|
9C94
 
 | | Crystal structure of menin in complex with inhibitor compound 20 | | Descriptor: | Menin, {5-fluoro-2-[(5-{7-[(1-methylcyclopropyl)methyl]-2,7-diazaspiro[3.5]nonan-2-yl}-1,2,4-triazin-6-yl)oxy]phenyl}[(1R,5S)-3-oxa-8-azabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Bester, S.M, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Design of Potent Menin-KMT2A Interaction Inhibitors with Improved In Vitro ADME Properties and Reduced hERG Affinity.
Acs Med.Chem.Lett., 16, 2025
|
|
9C55
 
 | | Crystal structure of human PTPN2 in complex with active site inhibitor | | Descriptor: | 5-(3-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}PHENYL)-4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Bester, S.M, Linwood, R, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-06-05 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Enhancing the apo protein tyrosine phosphatase non-receptor type 2 crystal soaking strategy through inhibitor-accessible binding sites.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
9C54
 
 | | Crystal structure of human PTPN2 catalytic domain | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Bester, S.M, Linwood, R, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-06-05 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enhancing the apo protein tyrosine phosphatase non-receptor type 2 crystal soaking strategy through inhibitor-accessible binding sites.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
