3P0J
 
 | |
3IAL
 
 | | Giardia lamblia Prolyl-tRNA synthetase in complex with prolyl-adenylate | | Descriptor: | 5'-O-[(R)-hydroxy{[(2S)-pyrrolidin-2-ylcarbonyl]oxy}phosphoryl]adenosine, GLYCEROL, Prolyl-tRNA synthetase | | Authors: | Larson, E.T, Merritt, E.A, Medical Structural Genomics of Pathogenic Protozoa, Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2009-07-14 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the prolyl-tRNA synthetase from the eukaryotic pathogen Giardia lamblia.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3K34
 
 | | Human carbonic anhydrase II with a sulfonamide inhibitor | | Descriptor: | (4-SULFAMOYL-PHENYL)-THIOCARBAMIC ACID O-(2-THIOPHEN-3-YL-ETHYL) ESTER, 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Behnke, C.A, Le Trong, I, Merritt, E.A, Teller, D.C, Stenkamp, R.E. | | Deposit date: | 2009-10-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic resolution studies of carbonic anhydrase II.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3KFL
 
 | | Leishmania major methionyl-tRNA synthetase in complex with methionyladenylate and pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Larson, E.T, Merritt, E.A, Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2009-10-27 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Leishmania major methionyl-tRNA synthetase in complex with intermediate products methionyladenylate and pyrophosphate.
Biochimie, 93, 2011
|
|
3KKK
 
 | |
3LSQ
 
 | |
1ITH
 
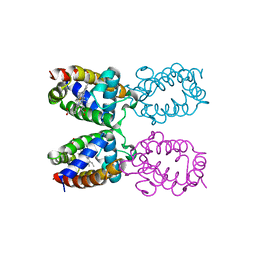 | | STRUCTURE DETERMINATION AND REFINEMENT OF HOMOTETRAMERIC HEMOGLOBIN FROM URECHIS CAUPO AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | CYANIDE ION, HEMOGLOBIN (CYANO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hackert, M, Kolatkar, P, Ernst, S.R, Ogata, C.M, Hendrickson, W.A, Merritt, E.A, Phizackerley, R.P. | | Deposit date: | 1991-12-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination and refinement of homotetrameric hemoglobin from Urechis caupo at 2.5 A resolution.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
1MK5
 
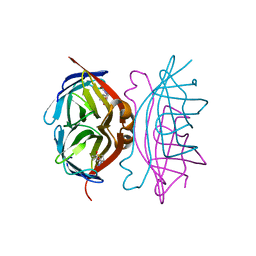 | | Wildtype Core-Streptavidin with Biotin at 1.4A. | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hyre, D.E, Le Trong, I, Merritt, E.A, Green, N.M, Stenkamp, R.E, Stayton, P.S. | | Deposit date: | 2002-08-28 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cooperative hydrogen bond interactions in the streptavidin-biotin system
Protein Sci., 15, 2006
|
|
1MD2
 
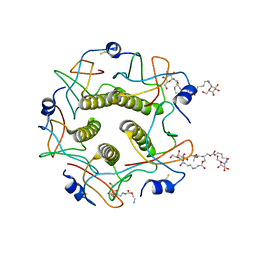 | | CHOLERA TOXIN B-PENTAMER WITH DECAVALENT LIGAND BMSC-0013 | | Descriptor: | 3-ETHYLAMINO-4-METHYLAMINO-CYCLOBUTANE-1,2-DIONE, CHOLERA TOXIN B SUBUNIT, CYANIDE ION, ... | | Authors: | Zhang, Z, Merritt, E.A, Ahn, M, Roach, C, Hol, W.G.J, Fan, E. | | Deposit date: | 2002-08-06 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Solution and crystallographic studies of branched multivalent ligands that inhibit the receptor-binding of cholera toxin.
J.Am.Chem.Soc., 124, 2002
|
|
1MEP
 
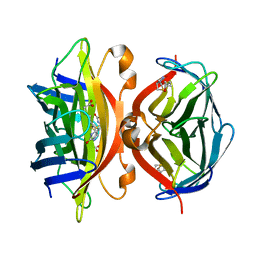 | | Crystal Structure of Streptavidin Double Mutant S45A/D128A with Biotin: Cooperative Hydrogen-Bond Interactions in the Streptavidin-Biotin System. | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hyre, D.E, Le Trong, I, Merritt, E.A, Stenkamp, R.E, Green, N.M, Stayton, P.S. | | Deposit date: | 2002-08-08 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cooperative hydrogen bond interactions in the streptavidin-biotin system
Protein Sci., 15, 2006
|
|
5SWH
 
 | | c-Src V281C kinase domain in complex with Rao-IV-151 | | Descriptor: | (2R)-3-[4-amino-5-(4-chlorophenyl)-7-(2-methoxyethyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl]-2-cyano-N-(propan-2-yl)propanami de, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Merritt, E.A, Dieter, E.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A combined approach reveals a regulatory mechanism coupling Src's kinase activity, localization, and phosphotransferase-independent functions
Mol.Cell, 2019
|
|
2F67
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.6 A resolution with BENZO[CD]INDOL-2(1H)-ONE bound | | Descriptor: | BENZO[CD]INDOL-2(1H)-ONE, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2F62
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.5 A resolution with (2-ETHYLPHENYL)METHANOL bound | | Descriptor: | (2-ETHYLPHENYL)METHANOL, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2F2T
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.7 A resolution with 5-Aminoisoquinoline bound | | Descriptor: | GLYCEROL, ISOQUINOLIN-5-AMINE, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-17 | | Release date: | 2005-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2F64
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.6 A resolution with 1-METHYLQUINOLIN-2(1H)-ONE bound | | Descriptor: | 1-METHYLQUINOLIN-2(1H)-ONE, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2A0K
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.8 A resolution | | Descriptor: | GLYCEROL, Nucleoside 2-deoxyribosyltransferase, SULFATE ION | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-06-16 | | Release date: | 2005-07-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2B4R
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 Angstrom Resolution reveals intriguing extra electron density in the active site | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Robien, M.A, Bosch, J, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 A resolution reveals intriguing extra electron density in the active site
Proteins, 62, 2006
|
|
2B4T
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 Angstrom resolution reveals intriguing extra electron density in the active site | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Robien, M.A, Bosch, J, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 A resolution reveals intriguing extra electron density in the active site
Proteins, 62, 2006
|
|
2CBP
 
 | | CUCUMBER BASIC PROTEIN, A BLUE COPPER PROTEIN | | Descriptor: | COPPER (II) ION, CUCUMBER BASIC PROTEIN | | Authors: | Guss, J.M, Freeman, H.C. | | Deposit date: | 1996-03-16 | | Release date: | 1997-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a phytocyanin, the basic blue protein from cucumber, refined at 1.8 A resolution.
J.Mol.Biol., 262, 1996
|
|
2GJF
 
 | |
1LT3
 
 | | HEAT-LABILE ENTEROTOXIN DOUBLE MUTANT N40C/G166C | | Descriptor: | HEAT-LABILE ENTEROTOXIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Van Den Akker, F, Hol, W.G.J. | | Deposit date: | 1997-04-12 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of heat-labile enterotoxin from Escherichia coli with increased thermostability introduced by an engineered disulfide bond in the A subunit.
Protein Sci., 6, 1997
|
|
1LTG
 
 | |
5T0P
 
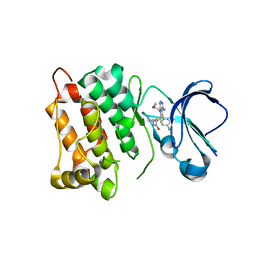 | | c-Src kinase domain in complex with Rao-IV-151 | | Descriptor: | (2E)-3-[4-amino-5-(4-chlorophenyl)-7-(2-methoxyethyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl]-2-cyano-N-(propan-2-yl)prop-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Dieter, E.M, Merritt, E.A, Maly, D.J. | | Deposit date: | 2016-08-16 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A chemical genetic method for conformation-selective kinase inhibition
To Be Published
|
|
5SYS
 
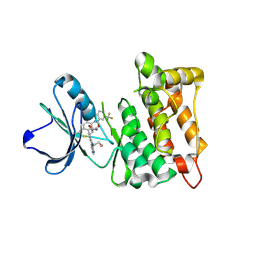 | | c-Src V281C bound to N-[3-({6-[(1E)-2-cyano-3-(methylamino)-3-oxoprop-1-en-1-yl]-7-(2-methoxyethyl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl}ethynyl)-4-methylphenyl]-3-(trifluoromethyl)benzamide inhibitor | | Descriptor: | N-[3-({6-[(2S)-2-cyano-3-(methylamino)-3-oxopropyl]-7-(2-methoxyethyl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl}ethynyl)-4-methylphenyl]-3-(trifluoromethyl)benzamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Dieter, E.M, Merritt, E.A, Maly, D.J. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A combined approach reveals a regulatory mechanism coupling Src's kinase activity, localization, and phosphotransferase-independent functions
Mol.Cell, 2019
|
|
5TEH
 
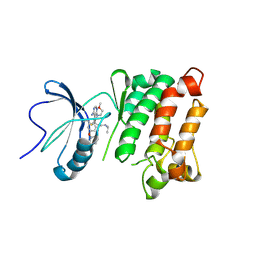 | | c-Src V281C kinase domain in complex with Rao-IV-156 | | Descriptor: | (2S)-3-[4-amino-7-(2-methoxyethyl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl]-2-cyano-N-(propan-2-yl)propanamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Dieter, E.M, Merritt, E.A, Maly, D.J. | | Deposit date: | 2016-09-21 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | A combined approach reveals a regulatory mechanism coupling Src's kinase activity, localization, and phosphotransferase-independent functions
Mol.Cell, 2019
|
|
