8ZB5
 
 | | Crystal structure of NudC from Mycobacterium abscessus in complex with AMP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Meng, L, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2024-04-26 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies on Mycobacterial NudC Reveal a Class of Zinc Independent NADH Pyrophosphatase.
J.Mol.Biol., 436, 2024
|
|
8ZB4
 
 | | Crystal structure of NudC from Mycobacterium abscessus in complex with NAD | | Descriptor: | CALCIUM ION, NAD(+) diphosphatase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Meng, L, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2024-04-26 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Studies on Mycobacterial NudC Reveal a Class of Zinc Independent NADH Pyrophosphatase.
J.Mol.Biol., 436, 2024
|
|
8ZB3
 
 | | Crystal structure of NudC from Mycobacterium abscessus | | Descriptor: | CALCIUM ION, IMIDAZOLE, NAD(+) diphosphatase | | Authors: | Meng, L, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2024-04-26 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies on Mycobacterial NudC Reveal a Class of Zinc Independent NADH Pyrophosphatase.
J.Mol.Biol., 436, 2024
|
|
5XA6
 
 | |
4MPS
 
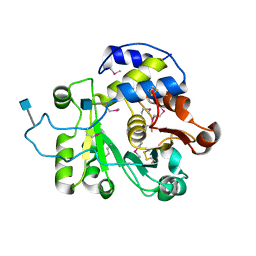 | | Crystal structure of rat Beta-galactoside alpha-2,6-sialyltransferase 1 (ST6GAL1), Northeast Structural Genomics Consortium Target RnR367A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactoside alpha-2,6-sialyltransferase 1 | | Authors: | Forouhar, F, Meng, L, Milaninia, S, Seetharaman, J, Su, M, Kornhaber, G, Montelione, G.T, Hunt, J.F, Moremen, K.W, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-09-13 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzymatic Basis for N-Glycan Sialylation: STRUCTURE OF RAT alpha 2,6-SIALYLTRANSFERASE (ST6GAL1) REVEALS CONSERVED AND UNIQUE FEATURES FOR GLYCAN SIALYLATION.
J.Biol.Chem., 288, 2013
|
|
7WQV
 
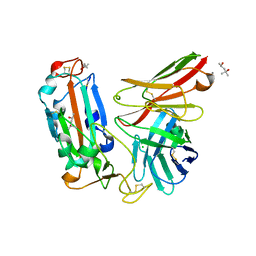 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | Authors: | Zha, J, Meng, L, Zhang, X, Li, D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
1WKM
 
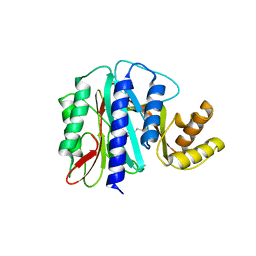 | | THE PRODUCT BOUND FORM OF THE MN(II)LOADED METHIONINE AMINOPEPTIDASE FROM HYPERTHERMOPHILE PYROCOCCUS FURIOSUS | | Descriptor: | MANGANESE (II) ION, METHIONINE, Methionine aminopeptidase | | Authors: | Copik, A.J, Nocek, B.P, Jang, S.B, Swierczek, S.I, Ruebush, S, Meng, L, D'souza, V.M, Peters, J.W, Bennett, B, Holz, R.C. | | Deposit date: | 2004-06-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | EPR and X-ray crystallographic characterization of the product-bound form of the MnII-loaded methionyl aminopeptidase from Pyrococcus furiosus
Biochemistry, 44, 2005
|
|
2KUO
 
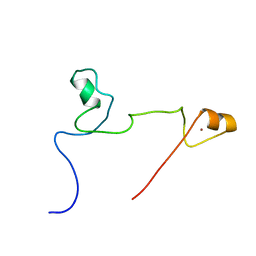 | | Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Li, G.Y, McCulloch, R.D, Fenton, A, Cheung, M, Meng, L, Ikura, M, Koch, C.A. | | Deposit date: | 2010-02-23 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and identification of ADP-ribose recognition motifs of aprataxin PNK-like factor (APLF) required for the interaction with sites of DNA damage response
To be Published
|
|
5BR8
 
 | | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sierra, R.G, Gati, C, Laksmono, H, Dao, E.H, Gul, S, Fuller, F, Kern, J, Chatterjee, R, Ibrahim, M, Brewster, A, Young, I.D, Michels-Clark, T, Aquila, A, Mengning, L, Hunter, M.S, Koglin, J.E, Boutet, S, Junco, E.A, Hayes, B, Bogan, M.J, Hampton, C.Y, Puglisi, E.V, Sauter, N.K, Stan, C.A, Zouni, A, Yano, J, Yachandra, V.K, Soltis, S.M, Puglisi, J.D, DeMirci, H. | | Deposit date: | 2015-05-29 | | Release date: | 2015-11-18 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin
To Be Published
|
|
6CSV
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 63 kDa,Centrosomal protein of 152 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
6CSU
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 152 kDa, Centrosomal protein of 63 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
7QEL
 
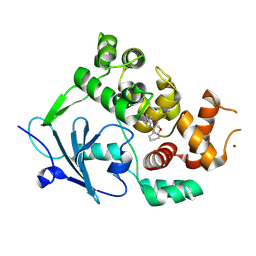 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011247 | | Descriptor: | (2~{R})-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)spiro[2.3]hexane-2-carboxamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Virtual fragment screening for DNA repair inhibitors in vast chemical space.
Nat Commun, 16, 2025
|
|
6N45
 
 | |
7MXD
 
 | |
7N28
 
 | |
5B6K
 
 | |
8CEY
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH11233. | | Descriptor: | N-glycosylase/DNA lyase, NICKEL (II) ION, ~{N}-(3,3-dimethylcyclobutyl)imidazo[5,1-b][1,3]thiazole-3-carboxamide | | Authors: | Kosenina, S, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Virtual fragment screening for DNA repair inhibitors in vast chemical space.
Nat Commun, 16, 2025
|
|
8CEX
 
 | |
6APL
 
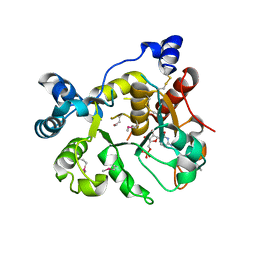 | | Crystal Structure of human ST6GALNAC2 in complex with CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Forouhar, F, Moremen, K.W, Northeast Structural Genomics Consortium (NESG), Tong, L. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Expression system for structural and functional studies of human glycosylation enzymes.
Nat. Chem. Biol., 14, 2018
|
|
6APJ
 
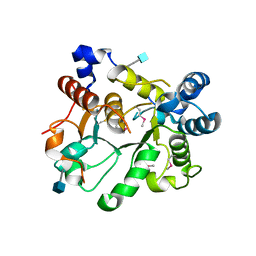 | | Crystal Structure of human ST6GALNAC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 | | Authors: | Forouhar, F, Moremen, K.W, Northeast Structural Genomics Consortium (NESG), Tong, L. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Expression system for structural and functional studies of human glycosylation enzymes.
Nat. Chem. Biol., 14, 2018
|
|
5XGH
 
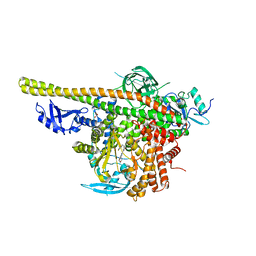 | | Crystal structure of PI3K complex with an inhibitor | | Descriptor: | 3-[(4-fluorophenyl)methylamino]-5-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Song, K, Yang, X, Zhao, Y, Jian, Z. | | Deposit date: | 2017-04-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | New Insights into PI3K Inhibitor Design using X-ray Structures of PI3K alpha Complexed with a Potent Lead Compound.
Sci Rep, 7, 2017
|
|
4L23
 
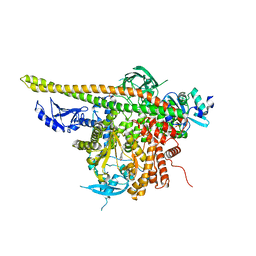 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha and PI-103 | | Descriptor: | 3-(4-MORPHOLIN-4-YLPYRIDO[3',2':4,5]FURO[3,2-D]PYRIMIDIN-2-YL)PHENOL, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-04 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
4L2Y
 
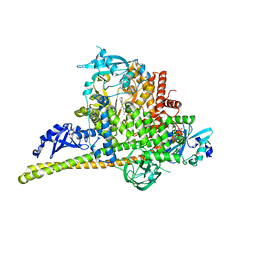 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha and compound 9d | | Descriptor: | 3-amino-5-[4-(morpholin-4-yl)pyrido[3',2':4,5]furo[3,2-d]pyrimidin-2-yl]phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
4L1B
 
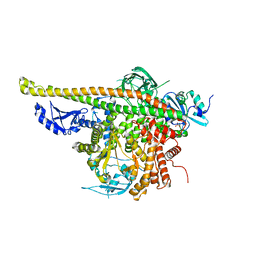 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-03 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
7W91
 
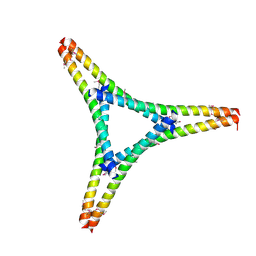 | | Residues 440-490 of centrosomal protein 63 | | Descriptor: | Centrosomal protein of 63 kDa | | Authors: | Yun, H.Y, Ku, B. | | Deposit date: | 2021-12-09 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.292 Å) | | Cite: | Architectural basis for cylindrical self-assembly governing Plk4-mediated centriole duplication in human cells.
Commun Biol, 6, 2023
|
|
