1KLQ
 
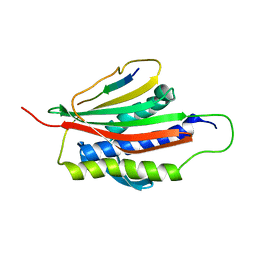 | | The Mad2 Spindle Checkpoint Protein Undergoes Similar Major Conformational Changes upon Binding to Either Mad1 or Cdc20 | | Descriptor: | MITOTIC SPINDLE ASSEMBLY CHECKPOINT PROTEIN MAD2A, Mad2-binding peptide | | Authors: | Luo, X, Tang, Z, Rizo, J, Yu, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-01-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20.
Mol.Cell, 9, 2002
|
|
4ZUZ
 
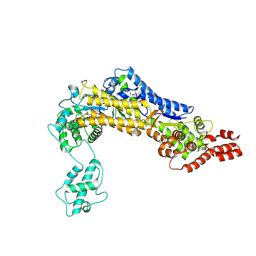 | | SidC 1-871 | | Descriptor: | SidC | | Authors: | Luo, X, Wasilko, D.J, Liu, Y, Sun, J, Wu, X, Luo, Z.-Q, Mao, Y. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of the Legionella Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome.
Plos Pathog., 11, 2015
|
|
1S2H
 
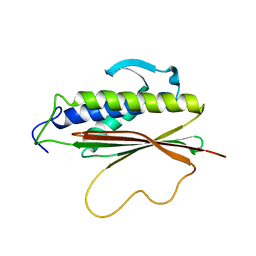 | | The Mad2 spindle checkpoint protein possesses two distinct natively folded states | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A | | Authors: | Luo, X, Tang, Z, Xia, G, Wassmann, K, Matsumoto, T, Rizo, J, Yu, H. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein has two distinct natively folded states.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1DUJ
 
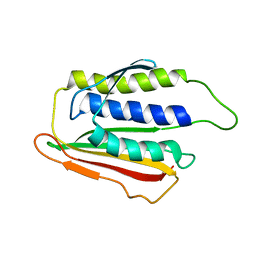 | | SOLUTION STRUCTURE OF THE SPINDLE ASSEMBLY CHECKPOINT PROTEIN HUMAN MAD2 | | Descriptor: | SPINDLE ASSEMBLY CHECKPOINT PROTEIN | | Authors: | Luo, X, Fang, G, Coldiron, M, Lin, Y, Yu, H. | | Deposit date: | 2000-01-17 | | Release date: | 2000-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20.
Nat.Struct.Biol., 7, 2000
|
|
1TBD
 
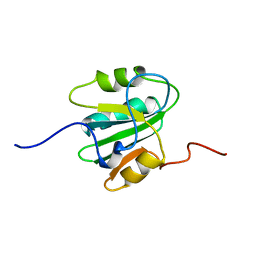 | | SOLUTION STRUCTURE OF THE ORIGIN DNA BINDING DOMAIN OF SV40 T-ANTIGEN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SV40 T-ANTIGEN | | Authors: | Luo, X, Sanford, D.G, Bullock, P.A, Bachovchin, W.W. | | Deposit date: | 1996-11-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the origin DNA-binding domain of SV40 T-antigen.
Nat.Struct.Biol., 3, 1996
|
|
5U36
 
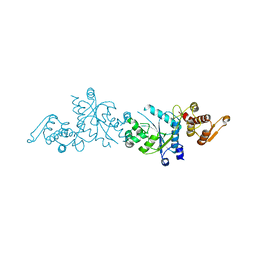 | | Crystal Structure Of A Mutant M. Jannashii Tyrosyl-tRNA Synthetase | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Luo, X, Fu, G, Zhu, X, Wilson, I.A, Wang, F. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria.
Nat. Chem. Biol., 13, 2017
|
|
3D3C
 
 | |
3D3B
 
 | |
2TBD
 
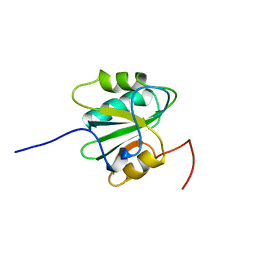 | | SV40 T ANTIGEN DNA-BINDING DOMAIN, NMR, 30 STRUCTURES | | Descriptor: | SV40 T ANTIGEN | | Authors: | Luo, X, Sanford, D.G, Bullock, P.A, Bachovchin, W.W. | | Deposit date: | 1997-01-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the origin DNA-binding domain of SV40 T-antigen.
Nat.Struct.Biol., 3, 1996
|
|
6L1W
 
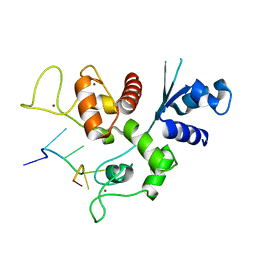 | | Zinc-finger Antiviral Protein (ZAP) bound to RNA | | Descriptor: | RNA (5'-R(*CP*GP*UP*CP*GP*U)-3'), ZINC ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Luo, X, Wang, X, Gao, Y, Zhu, J, Liu, S, Gao, G, Gao, P. | | Deposit date: | 2019-09-30 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Molecular Mechanism of RNA Recognition by Zinc-Finger Antiviral Protein.
Cell Rep, 30, 2020
|
|
4N14
 
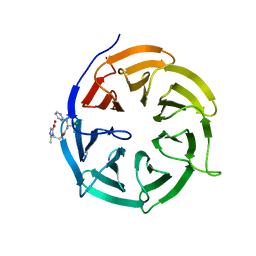 | | Crystal structure of Cdc20 and apcin complex | | Descriptor: | 2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl [(1R)-2,2,2-trichloro-1-(pyrimidin-2-ylamino)ethyl]carbamate, Cell division cycle protein 20 homolog | | Authors: | Luo, X, Tian, W, Yu, H. | | Deposit date: | 2013-10-03 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C.
Nature, 514, 2014
|
|
4DZO
 
 | |
5HGU
 
 | |
5BRM
 
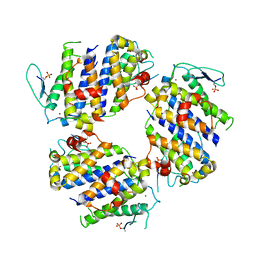 | |
3IMQ
 
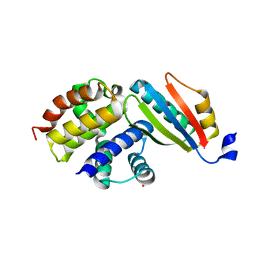 | | Crystal structure of the NusB101-S10(delta loop) complex | | Descriptor: | 30S ribosomal protein S10, N utilization substance protein B, POTASSIUM ION | | Authors: | Luo, X, Wahl, M.C. | | Deposit date: | 2009-08-11 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fine tuning of the E. coli NusB:NusE complex affinity to BoxA RNA is required for processive antitermination.
Nucleic Acids Res., 38, 2010
|
|
4GGD
 
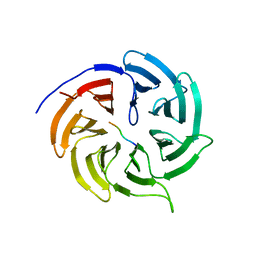 | |
4GGA
 
 | |
4GGC
 
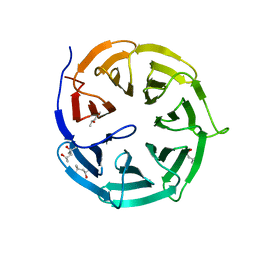 | |
4LGD
 
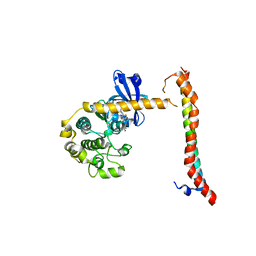 | | Structural Basis for Autoactivation of Human Mst2 Kinase and Its Regulation by RASSF5 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ras association domain family member 5, ... | | Authors: | Luo, X, Ni, L, Tomchick, D.R. | | Deposit date: | 2013-06-27 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Basis for Autoactivation of Human Mst2 Kinase and Its Regulation by RASSF5.
Structure, 21, 2013
|
|
4LG4
 
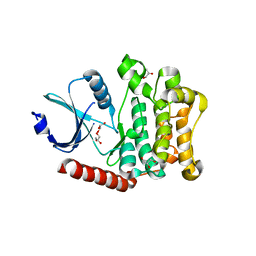 | |
5XPI
 
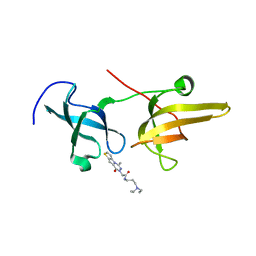 | | Structure of UHRF1 TTD in complex with NV01 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, N-[3-(diethylamino)propyl]-2-(12-methyl-9-oxidanylidene-5-thia-1,10,11-triazatricyclo[6.4.0.0^2,6]dodeca-2(6),3,7,11-tetraen-10-yl)ethanamide | | Authors: | Luo, X, Zhao, K. | | Deposit date: | 2017-06-02 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Small-Molecule Antagonists of the H3K9me3 Binding to UHRF1 Tandem Tudor Domain
SLAS Discov, 23, 2018
|
|
3L15
 
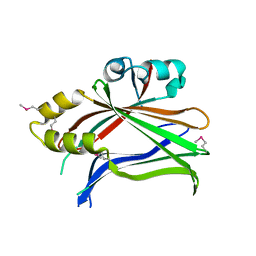 | | Human Tead2 transcriptional factor | | Descriptor: | GLYCEROL, Transcriptional enhancer factor TEF-4 | | Authors: | Tomchick, D.R, Luo, X, Tian, W. | | Deposit date: | 2009-12-10 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the YAP-binding domain of human TEAD2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7QK4
 
 | | EED in complex with PRC2 allosteric inhibitor compound 22 (MAK683) | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methyl]-8-(2-methylpyridin-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QJU
 
 | | EED in complex with PRC2 allosteric inhibitor compound 7 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QJG
 
 | | EED in complex with PRC2 allosteric inhibitor compound 6 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-7-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
