4TJX
 
 | | Crystal structure of protease-associated domain of Arabidopsis VSR1 in complex with aleurain peptide | | 分子名称: | Aleurain peptide, Vacuolar-sorting receptor 1 | | 著者 | Luo, F, Fong, Y.H, Jiang, L.W, Wong, K.B. | | 登録日 | 2014-05-25 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | How vacuolar sorting receptor proteins interact with their cargo proteins: crystal structures of apo and cargo-bound forms of the protease-associated domain from an Arabidopsis vacuolar sorting receptor.
Plant Cell, 26, 2014
|
|
4TJV
 
 | | Crystal structure of protease-associated domain of Arabidopsis vacuolar sorting receptor 1 | | 分子名称: | IODIDE ION, Vacuolar-sorting receptor 1 | | 著者 | Luo, F, Fong, Y.H, Jiang, L.W, Wong, K.B. | | 登録日 | 2014-05-25 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | How vacuolar sorting receptor proteins interact with their cargo proteins: crystal structures of apo and cargo-bound forms of the protease-associated domain from an Arabidopsis vacuolar sorting receptor.
Plant Cell, 26, 2014
|
|
5XSG
 
 | | Ultrahigh resolution structure of FUS (37-42) SYSGYS determined by MicroED | | 分子名称: | RNA-binding protein FUS | | 著者 | Luo, F, Gui, X, Zhou, H, Li, D, Li, X, Liu, C. | | 登録日 | 2017-06-14 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.73 Å) | | 主引用文献 | Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7EMN
 
 | |
6J60
 
 | | hnRNP A1 reversible amyloid core GFGGNDNFG (residues 209-217) | | 分子名称: | 9-mer peptide (GFGGNDNFG) from Heterogeneous nuclear ribonucleoprotein A1 | | 著者 | Luo, F, Zhou, H, Gui, X, Li, D, Li, X, Liu, C. | | 登録日 | 2019-01-12 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.96 Å) | | 主引用文献 | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
2M01
 
 | | Solution structure of Kunitz-type neurotoxin LmKKT-1a from scorpion venom | | 分子名称: | Protease inhibitor LmKTT-1a | | 著者 | Luo, F, Jiang, L, Liu, M, Chen, Z, Wu, Y. | | 登録日 | 2012-10-15 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Genomic and structural characterization of Kunitz-type peptide LmKTT-1a highlights diversity and evolution of scorpion potassium channel toxins.
Plos One, 8, 2013
|
|
1EV7
 
 | | CRYSTAL STRUCTURE OF DNA RESTRICTION ENDONUCLEASE NAEI | | 分子名称: | TYPE IIE RESTRICTION ENDONUCLEASE NAEI | | 著者 | Huai, Q, Colandene, J.D, Chen, Y, Luo, F, Zhao, Y. | | 登録日 | 2000-04-19 | | 公開日 | 2000-10-19 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Crystal structure of NaeI-an evolutionary bridge between DNA endonuclease and topoisomerase.
EMBO J., 19, 2000
|
|
6A6B
 
 | | cryo-em structure of alpha-synuclein fiber | | 分子名称: | Alpha-synuclein | | 著者 | Li, Y.W, Zhao, C.Y, Luo, F, Liu, Z, Gui, X, Luo, Z, Zhang, X, Li, D, Liu, C, Li, X. | | 登録日 | 2018-06-27 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy
Cell Res., 28, 2018
|
|
6KJ3
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.60 A | | 分子名称: | RNA-binding protein FUS | | 著者 | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | 登録日 | 2019-07-20 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.6 Å) | | 主引用文献 | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KJ4
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.65 A | | 分子名称: | RNA-binding protein FUS | | 著者 | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | 登録日 | 2019-07-20 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.65 Å) | | 主引用文献 | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KJ2
 
 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.67 A | | 分子名称: | RNA-binding protein FUS | | 著者 | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | 登録日 | 2019-07-20 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.67 Å) | | 主引用文献 | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KJ1
 
 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.65 A | | 分子名称: | RNA-binding protein FUS | | 著者 | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | 登録日 | 2019-07-20 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.65 Å) | | 主引用文献 | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
5ZGD
 
 | | hnRNPA1 reversible amyloid core GFGGNDNFG (residues 209-217) determined by X-ray | | 分子名称: | GLY-PHE-GLY-GLY-ASN-ASP-ASN-PHE-GLY | | 著者 | Gui, X, Xie, M, Zhao, M, Luo, F, He, J, Li, D, Liu, C. | | 登録日 | 2018-03-08 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.401 Å) | | 主引用文献 | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
5ZGL
 
 | | hnRNP A1 segment GGGYGGS (residues 234-240) | | 分子名称: | 7-mer peptide from Heterogeneous nuclear ribonucleoprotein A1 | | 著者 | Xie, M, Luo, F, Gui, X, Zhao, M, He, J, Li, D, Liu, C. | | 登録日 | 2018-03-09 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
7C4S
 
 | | Sphingosine-1-phosphate receptor 3 with a natural ligand. | | 分子名称: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Antibody Fab fragment heavy chain, Antibody Fab fragment light chain, ... | | 著者 | Iwata, S, Maeda, S, Luo, F, Nango, E, hirata, K, Asada, H. | | 登録日 | 2020-05-18 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Endogenous agonist-bound S1PR3 structure reveals determinants of G protein-subtype bias.
Sci Adv, 7, 2021
|
|
7DGE
 
 | | intermediate state of class C GPCR | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 1, nanobody | | 著者 | Zhang, J.Y, Wu, L.J, Luo, F, Hua, T, Liu, Z.J. | | 登録日 | 2020-11-11 | | 公開日 | 2021-09-22 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Structural insights into the activation initiation of full-length mGlu1.
Protein Cell, 12, 2021
|
|
7DGD
 
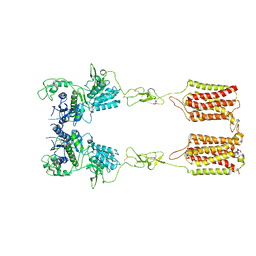 | | apo state of class C GPCR | | 分子名称: | Metabotropic glutamate receptor 1 | | 著者 | Zhang, J.Y, Wu, L.J, Luo, F, Hua, T, Liu, Z.J. | | 登録日 | 2020-11-11 | | 公開日 | 2021-09-22 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Structural insights into the activation initiation of full-length mGlu1.
Protein Cell, 12, 2021
|
|
7Y3G
 
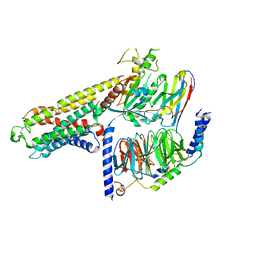 | | Cryo-EM structure of a class A orphan GPCR | | 分子名称: | G-protein coupled receptor 12, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Liu, Z.J, Hua, T, Li, H, Zhang, J.Y, Luo, F. | | 登録日 | 2022-06-10 | | 公開日 | 2023-06-07 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structural insight into the constitutive activity of human orphan receptor GPR12.
Sci Bull (Beijing), 68, 2023
|
|
2RMC
 
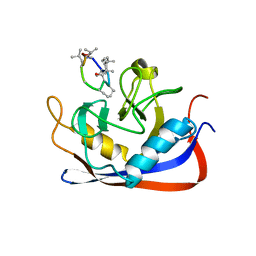 | | Crystal structure of murine cyclophilin C complexed with immunosuppressive drug cyclosporin A | | 分子名称: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE C | | 著者 | Ke, H, Zhao, Y, Luo, F, Weissman, I, Friedman, J. | | 登録日 | 1994-01-07 | | 公開日 | 1995-02-14 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal Structure of Murine Cyclophilin C Complexed with Immunosuppressive Drug Cyclosporin A
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
5XDQ
 
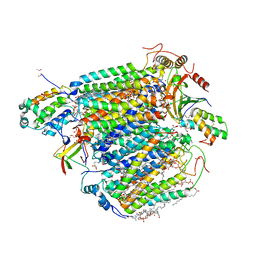 | | Bovine heart cytochrome c oxidase in the fully oxidized state with pH 7.3 at 1.77 angstrom resolution | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | 著者 | Luo, F.J, Shimada, A, Hagimoto, N, Shimada, S, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | 登録日 | 2017-03-29 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structure of bovine cytochrome c oxidase crystallized at a neutral pH using a fluorinated detergent.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5XDX
 
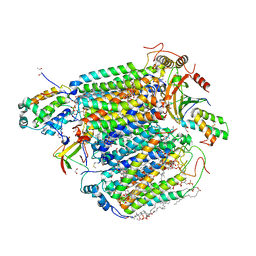 | | Bovine heart cytochrome c oxidase in the reduced state with pH 7.3 at 1.99 angstrom resolution | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | 著者 | Luo, F.J, Shimada, A, Hagimoto, N, Shimada, S, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | 登録日 | 2017-03-30 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure of bovine cytochrome c oxidase in the ligand-free reduced state at neutral pH.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5XRR
 
 | | Crystal structure of FUS (54-59) SYSSYG | | 分子名称: | RNA-binding protein FUS, ZINC ION | | 著者 | Zhao, M, Gui, X, Li, D, Liu, C. | | 登録日 | 2017-06-09 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7QK4
 
 | | EED in complex with PRC2 allosteric inhibitor compound 22 (MAK683) | | 分子名称: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methyl]-8-(2-methylpyridin-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | 著者 | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | 登録日 | 2021-12-17 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.602 Å) | | 主引用文献 | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QJG
 
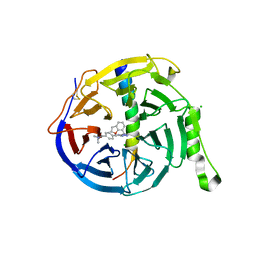 | | EED in complex with PRC2 allosteric inhibitor compound 6 | | 分子名称: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-7-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | 著者 | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | 登録日 | 2021-12-16 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QJU
 
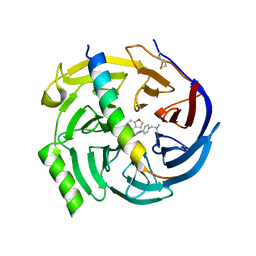 | | EED in complex with PRC2 allosteric inhibitor compound 7 | | 分子名称: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | 著者 | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | 登録日 | 2021-12-17 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
