8GYD
 
 | | Structure of Schistosoma japonicum Glutathione S-transferase bound with the ligand complex of 16 | | Descriptor: | (2R)-2-[[2-(5-chloranylthiophen-2-yl)-4-oxidanylidene-6-[2-(1H-1,2,3,4-tetrazol-5-yl)phenyl]quinazolin-3-yl]methyl]-3-(4-chlorophenyl)propanoic acid, ETHANOL, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Wen, X, Jin, R, Hu, H, Zhu, J, Song, W, Lu, X. | | Deposit date: | 2022-09-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery, SAR Study of GST Inhibitors from a Novel Quinazolin-4(1 H )-one Focused DNA-Encoded Library.
J.Med.Chem., 66, 2023
|
|
4WUU
 
 | | Structure of ESK1 in complex with HLA-A*0201/WT1 | | Descriptor: | ARG-MET-PHE-PRO-ASN-ALA-PRO-TYR-LEU, Beta-2-microglobulin, ESK1, ... | | Authors: | Ataie, N.J, Ng, H.L. | | Deposit date: | 2014-11-03 | | Release date: | 2015-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.047 Å) | | Cite: | Structure of a TCR-Mimic Antibody with Target Predicts Pharmacogenetics.
J.Mol.Biol., 428, 2016
|
|
6TFM
 
 | | Frizzled8 CRD | | Descriptor: | Frizzled-8 | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Antiepileptic Drug Carbamazepine Binds to a Novel Pocket on the Wnt Receptor Frizzled-8.
J.Med.Chem., 63, 2020
|
|
6TFB
 
 | | Carbamazepine binds Frizzled8 | | Descriptor: | Frizzled-8, GLYCEROL, benzo[b][1]benzazepine-11-carboxamide | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-11-13 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Antiepileptic Drug Carbamazepine Binds to a Novel Pocket on the Wnt Receptor Frizzled-8.
J.Med.Chem., 63, 2020
|
|
6TR5
 
 | | Melatonin-Notum complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | Structural characterization of melatonin as an inhibitor of the Wnt deacylase Notum.
J. Pineal Res., 68, 2020
|
|
6TR7
 
 | | N-[2-(5-fluoro-1H-indol-3-yl)ethyl]acetamide-Notum complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural characterization of melatonin as an inhibitor of the Wnt deacylase Notum.
J. Pineal Res., 68, 2020
|
|
6TR6
 
 | | N-acetylserotonin-Notum complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of melatonin as an inhibitor of the Wnt deacylase Notum.
J. Pineal Res., 68, 2020
|
|
6U4J
 
 | | Crystal structure of IDH1 R132H mutant in complex with FT-2102 | | Descriptor: | 5-{[(1S)-1-(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)ethyl]amino}-1-methyl-6-oxo-1,6-dihydropyridine-2-carbonitrile, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Toms, A.V, Lin, J. | | Deposit date: | 2019-08-25 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Based Design and Identification of FT-2102 (Olutasidenib), a Potent Mutant-Selective IDH1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
4Z55
 
 | |
4RXZ
 
 | |
5CUJ
 
 | | Crystal structure of Human Defensin-5 Y27A mutant crystal form 2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Human Enteric alpha-Defensin 5 Promotes Shigella Infection by Enhancing Bacterial Adhesion and Invasion.
Immunity, 2018
|
|
5CUI
 
 | | Crystal structure of Human Defensin-5 R28A mutant. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, CHLORIDE ION, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Human Enteric alpha-Defensin 5 Promotes Shigella Infection by Enhancing Bacterial Adhesion and Invasion.
Immunity, 2018
|
|
5CUM
 
 | |
6WYS
 
 | | Lon protease proteolytic domain | | Descriptor: | Lon protease homolog, mitochondrial, SULFATE ION | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2020-05-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.229 Å) | | Cite: | Structure-Based Design of Selective LONP1 Inhibitors for Probing In Vitro Biology.
J.Med.Chem., 64, 2021
|
|
6X1M
 
 | |
6WZV
 
 | | Lon protease proteolytic domain | | Descriptor: | Lon protease homolog, mitochondrial, N-[(1R)-1-borono-3-methylbutyl]-Nalpha-(pyrazine-2-carbonyl)-D-phenylalaninamide, ... | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2020-05-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-Based Design of Selective LONP1 Inhibitors for Probing In Vitro Biology.
J.Med.Chem., 64, 2021
|
|
6X27
 
 | |
6DMQ
 
 | |
6DMM
 
 | | Crystal structure of the G23A mutant of human alpha defensin HNP4. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Neutrophil defensin 4, SULFATE ION | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-06-05 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Systematic mutational analysis of human neutrophil alpha-defensin HNP4.
Biochim Biophys Acta Biomembr, 1861, 2019
|
|
6GBI
 
 | | Wnt signalling | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ly6/PLAUR domain-containing protein 6, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2018-04-14 | | Release date: | 2018-08-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of the Wnt signaling enhancer LYPD6 and its interactions with the Wnt coreceptor LRP6.
FEBS Lett., 592, 2018
|
|
1QQL
 
 | | THE CRYSTAL STRUCTURE OF FIBROBLAST GROWTH FACTOR 7/1 CHIMERA | | Descriptor: | FIBROBLAST GROWTH FACTOR 7/1 CHIMERA | | Authors: | Ye, S, Luo, Y, Pelletier, H, McKeehan, W.L. | | Deposit date: | 1999-06-07 | | Release date: | 2000-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs.
Biochemistry, 40, 2001
|
|
1QQK
 
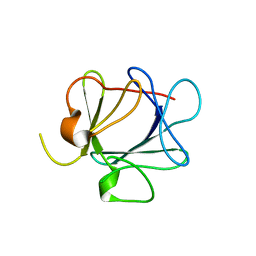 | | THE CRYSTAL STRUCTURE OF FIBROBLAST GROWTH FACTOR 7 (KERATINOCYTE GROWTH FACTOR) | | Descriptor: | FIBROBLAST GROWTH FACTOR 7 | | Authors: | Ye, S, Luo, Y, Pelletier, H, McKeehan, W.L. | | Deposit date: | 1999-06-07 | | Release date: | 2000-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs.
Biochemistry, 40, 2001
|
|
7VD4
 
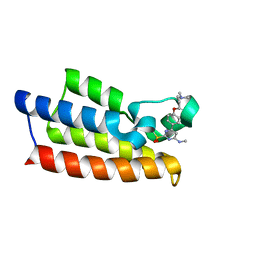 | | Crystal structure of BPTF-BRD with ligand TP248 bound | | Descriptor: | 6-[4-[3-(dimethylamino)propoxy]phenyl]-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B. | | Deposit date: | 2021-09-06 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85659146 Å) | | Cite: | Discovery of a highly potent CECR2 bromodomain inhibitor with 7H-pyrrolo[2,3-d] pyrimidine scaffold.
Bioorg.Chem., 123, 2022
|
|
3T2F
 
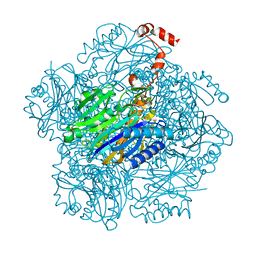 | | Fructose-1,6-bisphosphate aldolase/phosphatase from Thermoproteus neutrophilus, soaked with EDTA and DHAP | | Descriptor: | Fructose-1,6-bisphosphate aldolase/phosphatase, MAGNESIUM ION | | Authors: | Du, J, Say, R, Lue, W, Fuchs, G, Einsle, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase.
Nature, 478, 2011
|
|
3T2C
 
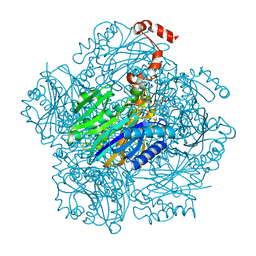 | | Fructose-1,6-bisphosphate aldolase/phosphatase from Thermoproteus neutrophilus, DHAP-bound form | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-1,6-bisphosphate aldolase/phosphatase, MAGNESIUM ION | | Authors: | Du, J, Say, R, Lue, W, Fuchs, G, Einsle, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase.
Nature, 478, 2011
|
|
