7S15
 
 | | GLP-1 receptor bound with Pfizer small molecule agonist | | Descriptor: | 2-[(4-{6-[(2,4-difluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-[(1-ethyl-1H-imidazol-5-yl)methyl]-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor | | Authors: | Liu, Y, Dias, J.M, Han, S. | | Deposit date: | 2021-09-01 | | Release date: | 2022-06-08 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Small-Molecule Oral Agonist of the Human Glucagon-like Peptide-1 Receptor.
J.Med.Chem., 65, 2022
|
|
5I5N
 
 | |
5I5O
 
 | |
4PEJ
 
 | | Crystal structure of a computationally designed retro-aldolase, RA110.4 (Cys free) | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
4PEK
 
 | | Crystal structure of a computationally designed retro-aldolase, RA114.3 | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
3BAR
 
 | | Crystal structure of Plasmodium falciparum orotidine 5'-phosphate decarboxylase covalently modified by 6-azido-UMP | | Descriptor: | Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Liu, Y, Bello, A.M, Poduch, E, Lau, W, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-11-08 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Relationships of C6-Uridine Derivatives Targeting Plasmodia Orotidine Monophosphate Decarboxylase.
J.Med.Chem., 51, 2008
|
|
4RZI
 
 | | Crystal structure of PhaB from Synechocystis sp. PCC 6803 | | Descriptor: | 3-ketoacyl-acyl carrier protein reductase | | Authors: | Xue, S, Liu, Y. | | Deposit date: | 2014-12-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.891 Å) | | Cite: | Structure-directed construction of a high-performance version of the enzyme FabG from the photosynthetic microorganism Synechocystis sp. PCC 6803.
Febs Lett., 589, 2015
|
|
4ZK7
 
 | | Crystal structure of rescued two-component self-assembling tetrahedral cage T33-31 | | Descriptor: | Chorismate mutase, Divalent-cation tolerance protein CutA | | Authors: | Liu, Y, Cascio, D, Sawaya, M.R, Bale, J, Collazo, M.J, Park, R, King, N, Baker, D, Yeates, T. | | Deposit date: | 2015-04-30 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a designed tetrahedral protein assembly variant engineered to have improved soluble expression.
Protein Sci., 24, 2015
|
|
2K5U
 
 | |
5EG8
 
 | | The cap binding site of influenza virus protein PB2 as a drug target | | Descriptor: | Polymerase basic protein 2, THIOCYANATE ION | | Authors: | Liu, Y, Zheng, X, Severin, C, Rocha de Moura, T, Li, K, Luo, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The cap-binding site of influenza virus protein PB2 as a drug target.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EQ0
 
 | | Crystal Structure of chromodomain of CBX8 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 8, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-12 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
5EPK
 
 | | Crystal Structure of chromodomain of CBX2 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 2, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
5EPL
 
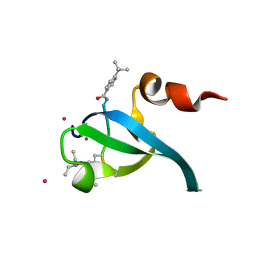 | | Crystal Structure of chromodomain of CBX4 in complex with inhibitor UNC3866 | | Descriptor: | E3 SUMO-protein ligase CBX4, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
3S9Y
 
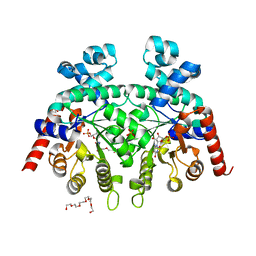 | | Crystal Structure of P. falciparum orotidine 5'-monophosphate decarboxylase complexed with 5-fluoro-6-amino-UMP in space group P21, produced from 5-fluoro-6-azido-UMP | | Descriptor: | 6-amino-5-fluorouridine 5'-(dihydrogen phosphate), DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Liu, Y, Kotra, L.P, Pai, E.F. | | Deposit date: | 2011-06-02 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents.
J.Med.Chem., 52, 2009
|
|
7XC5
 
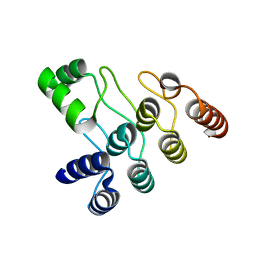 | | Crystal structure of the ANK domain of CLPB | | Descriptor: | Isoform 2 of Caseinolytic peptidase B protein homolog | | Authors: | Liu, Y, Wu, D, Lu, G, Gao, N, Lin, J. | | Deposit date: | 2022-03-23 | | Release date: | 2023-01-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization.
Plos Biol., 21, 2023
|
|
6ASG
 
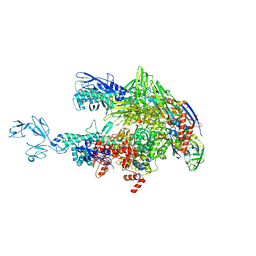 | | Crystal structure of Thermus thermophilus RNA polymerase core enzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, Y, Lin, W, Ying, R, Ebright, R.H. | | Deposit date: | 2017-08-24 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
2Q8Z
 
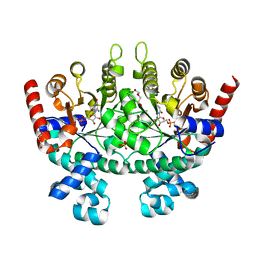 | | Crystal structure of Plasmodium falciparum orotidine 5'-phosphate decarboxylase complexed with 6-amino-UMP | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 6-AMINOURIDINE 5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Y, Lau, W, Bello, A.M, Kotra, L.P, Hui, R, Pai, E.F. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Activity Relationships of C6-Uridine Derivatives Targeting Plasmodia Orotidine Monophosphate Decarboxylase.
J.Med.Chem., 51, 2008
|
|
7WQR
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 28 | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ~{N}-(3-chlorophenyl)-2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]benzamide | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7WQS
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 25 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(3-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7WQM
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 24 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(2-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7EQ4
 
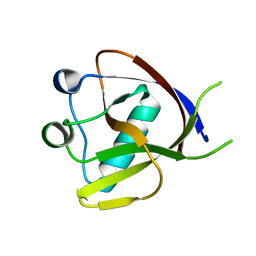 | | Crystal Structure of the N-terminus of Nonstructural protein 1 from SARS-CoV-2 | | Descriptor: | Host translation inhibitor nsp1 | | Authors: | Liu, Y, Ke, Z, Hu, H, Zhao, K, Xiao, J, Xia, Y, Li, Y. | | Deposit date: | 2021-04-29 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis and Function of the N Terminus of SARS-CoV-2 Nonstructural Protein 1.
Microbiol Spectr, 9, 2021
|
|
5ZBN
 
 | |
6GRB
 
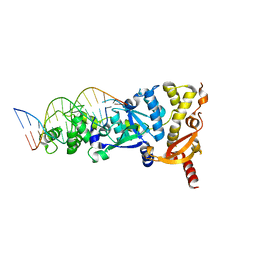 | | eukaryotic junction-resolving enzyme GEN-1 binding with Potassium | | Descriptor: | DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | Deposit date: | 2018-06-11 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
6C9K
 
 | | Single-Particle reconstruction of DARP14 - A designed protein scaffold displaying ~17kDa DARPin proteins | | Descriptor: | DARP14 - Subunit A with DARPin, DARP14 - Subunit B | | Authors: | Gonen, S, Liu, Y, Yeates, T.O, Gonen, T. | | Deposit date: | 2018-01-26 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Near-atomic cryo-EM imaging of a small protein displayed on a designed scaffolding system.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C9I
 
 | | Single-Particle reconstruction of DARP14 - A designed protein scaffold displaying ~17kDa DARPin proteins - Scaffold | | Descriptor: | DARP14 - Subunit A with DARPin, DARP14 - Subunit B | | Authors: | Gonen, S, Liu, Y, Yeates, T.O, Gonen, T. | | Deposit date: | 2018-01-26 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Near-atomic cryo-EM imaging of a small protein displayed on a designed scaffolding system.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
