2RHX
 
 | |
2RI3
 
 | |
2RHZ
 
 | |
8GRM
 
 | | Cryo-EM structure of PRC1 bound to H2AK119-UbcH5b-Ub nucleosome | | 分子名称: | COMMD3 protein, DNA (144-MER), DNA (145-MER), ... | | 著者 | Ai, H.S, Zebin, T, Zhihend, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | 登録日 | 2022-09-02 | | 公開日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
8GRQ
 
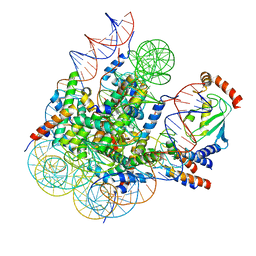 | | Cryo-EM structure of BRCA1/BARD1 bound to H2AK127-UbcH5c-Ub nucleosome | | 分子名称: | BRCA1-associated RING domain protein 1, Breast cancer type 1 susceptibility protein, DNA (147-MER), ... | | 著者 | Ai, H.S, Zebin, T, Zhiheng, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | 登録日 | 2022-09-02 | | 公開日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
8UCD
 
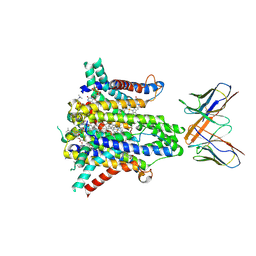 | | Cryo-EM structure of human STEAP1 in complex with AMG 509 Fab | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, AMG 509 anti-STEAP1 Fab, heavy chain, ... | | 著者 | Li, F, Bailis, J.M, Zhang, H. | | 登録日 | 2023-09-26 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | AMG 509 (Xaluritamig), an Anti-STEAP1 XmAb 2+1 T-cell Redirecting Immune Therapy with Avidity-Dependent Activity against Prostate Cancer.
Cancer Discov, 14, 2024
|
|
8HR1
 
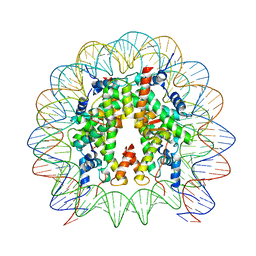 | | Cryo-EM structure of SSX1 bound to the unmodified nucleosome at a resolution of 3.02 angstrom | | 分子名称: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Zebin, T, Ai, H.S, Ziyu, X, Man, P, Liu, L. | | 登録日 | 2022-12-14 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HQY
 
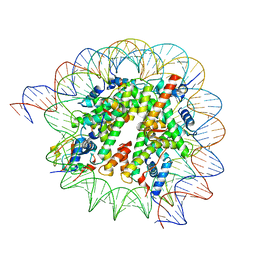 | | Cryo-EM structure of SSX1 bound to the H2AK119Ub nucleosome at a resolution of 3.05 angstrom | | 分子名称: | DNA (136-MER), DNA (137-MER), Histone H2A type 1-B/E, ... | | 著者 | Zebin, T, Ai, H.S, Ziyu, X, GuoChao, C, Man, P, Liu, L. | | 登録日 | 2022-12-14 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6IGO
 
 | |
6IGT
 
 | | MPZL1 mutant - V145G, Q146K, P147T and G148S | | 分子名称: | Myelin protein zero-like protein 1 | | 著者 | Yu, T. | | 登録日 | 2018-09-26 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.404 Å) | | 主引用文献 | Structural and biochemical studies of the extracellular domain of Myelin protein zero-like protein 1
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6IGW
 
 | | MPZL1 mutant - S86G, V145G, Q146K,P147T,G148S | | 分子名称: | Myelin protein zero-like protein 1 | | 著者 | Yu, T. | | 登録日 | 2018-09-26 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.979 Å) | | 主引用文献 | Structural and biochemical studies of the extracellular domain of Myelin protein zero-like protein 1
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6IWR
 
 | | Crystal structure of GalNAc-T7 with UDP, GalNAc and Mn2+ | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MANGANESE (II) ION, N-acetylgalactosaminyltransferase 7, ... | | 著者 | Yu, C, Yin, Y.X. | | 登録日 | 2018-12-06 | | 公開日 | 2019-02-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.604 Å) | | 主引用文献 | Structural basis of carbohydrate transfer activity of UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferase 7.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6IWQ
 
 | | Crystal structure of GalNAc-T7 with Mn2+ | | 分子名称: | MANGANESE (II) ION, N-acetylgalactosaminyltransferase 7 | | 著者 | Yu, C, Yin, Y.X. | | 登録日 | 2018-12-06 | | 公開日 | 2019-02-06 | | 最終更新日 | 2019-02-20 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural basis of carbohydrate transfer activity of UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferase 7.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6JXT
 
 | |
8GMT
 
 | | Structure of UmuD in complex with RecA filament | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), DNA polymerase V subunit UmuD, MAGNESIUM ION, ... | | 著者 | Gao, B, Feng, Y. | | 登録日 | 2022-08-22 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-01-18 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GMU
 
 | |
8GMS
 
 | | Structure of LexA in complex with RecA filament | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), LexA repressor, MAGNESIUM ION, ... | | 著者 | Gao, B, Feng, Y. | | 登録日 | 2022-08-22 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-01-18 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3G6L
 
 | |
3G6M
 
 | | crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with a potent inhibitor caffeine | | 分子名称: | CAFFEINE, Chitinase | | 著者 | Gan, Z, Yang, J, Lou, Z, Rao, Z, Zhang, K.-Q. | | 登録日 | 2009-02-06 | | 公開日 | 2010-02-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure and mutagenesis analysis of chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with the inhibitor caffeine
Microbiology, 156, 2010
|
|
6JX0
 
 | |
6JWL
 
 | |
6JX4
 
 | |
4TTH
 
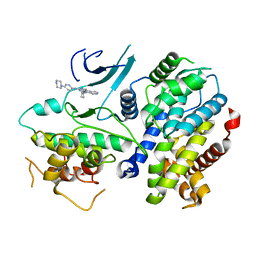 | | Crystal structure of a CDK6/Vcyclin complex with inhibitor bound | | 分子名称: | 9-cyclopentyl-N-(5-piperazin-1-ylpyridin-2-yl)pyrido[4,5]pyrrolo[1,2-d]pyrimidin-2-amine, Cyclin homolog, Cyclin-dependent kinase 6 | | 著者 | Piper, D.E, Walker, N, Wang, Z. | | 登録日 | 2014-06-20 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery of AMG 925, a FLT3 and CDK4 dual kinase inhibitor with preferential affinity for the activated state of FLT3.
J.Med.Chem., 57, 2014
|
|
7XCT
 
 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 2:1 complex | | 分子名称: | DNA (145-MER), Histone H2A, Histone H2B type 1-K, ... | | 著者 | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | 登録日 | 2022-03-25 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD1
 
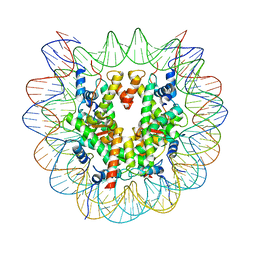 | | cryo-EM structure of unmodified nucleosome | | 分子名称: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | 登録日 | 2022-03-26 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
