8IDX
 
 | | Structure of p205 HIN | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Interferon-activable protein 205-B, ... | | Authors: | Li, Y.L, Jin, T.C. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of p205 HIN
to Be published
|
|
1NAV
 
 | | Thyroid Receptor Alpha in complex with an agonist selective for Thyroid Receptor Beta1 | | Descriptor: | SULFATE ION, hormone receptor alpha 1, THRA1, ... | | Authors: | Ye, L, Li, Y.L, Mellstrom, K, Mellin, C, Bladh, L.G, Koehler, K, Garg, N, Garcia Collazo, A.M, Litten, C, Husman, B, Persson, K, Ljunggren, J, Grover, G, Sleph, P.G, George, R, Malm, J. | | Deposit date: | 2002-11-29 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thyroid receptor ligands. 1. Agonist ligands selective for the thyroid receptor beta1.
J.Med.Chem., 46, 2003
|
|
1NAX
 
 | | Thyroid receptor beta1 in complex with a beta-selective ligand | | Descriptor: | Thyroid hormone receptor beta-1, {3,5-DICHLORO-4-[4-HYDROXY-3-(PROPAN-2-YL)PHENOXY]PHENYL}ACETIC ACID | | Authors: | Ye, L, Li, Y.L, Mellstrom, K, Mellin, C, Bladh, L.G, Koehler, K, Garg, N, Garcia Collazo, A.M, Litten, C, Husman, B, Persson, K, Ljunggren, J, Grover, G, Sleph, P.G, George, R, Malm, J. | | Deposit date: | 2002-11-29 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Thyroid receptor ligands. 1. Agonist ligands selective for the thyroid receptor beta1.
J.Med.Chem., 46, 2003
|
|
6J3X
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotriose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2019-01-06 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
6IYG
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotetraose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-15 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
6JI4
 
 | | brd4-bd1 bound with ligand 138 | | Descriptor: | (3R)-4-cyclopropyl-1,3-dimethyl-6-[5-methyl-4-(4-methylphenyl)-4H-1,2,4-triazol-3-yl]-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Cao, D.Y, Li, Y.L, Du, Z.Y, Li, J, Xiong, B. | | Deposit date: | 2019-02-20 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | brd4-bd1 bound with ligand 138
To Be Published
|
|
6JI5
 
 | | brd4-bd1 bound with ligand 167 | | Descriptor: | (3R)-4-cyclopentyl-6-[1-(2,4-dimethylphenyl)-3-(4-methylpiperazine-1-carbonyl)-1H-1,2,4-triazol-5-yl]-1,3-dimethyl-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Cao, D.Y, Li, Y.L, Du, Z.Y, Li, J, Xiong, B. | | Deposit date: | 2019-02-20 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | brd4-bd1 bound with ligand 167
To Be Published
|
|
6JI3
 
 | | BRD4-BD1 bound with ligand 103 | | Descriptor: | (3~{R})-4-cyclopropyl-1,3-dimethyl-6-(1~{H}-pyrrol-2-yl)-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Cao, D.Y, Li, Y.L, Du, Z.Y, Li, J, Xiong, B. | | Deposit date: | 2019-02-20 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | brd4-bd1 bound with ligand 103
To Be Published
|
|
6AIJ
 
 | | Cyclodextrin glycosyltransferase from Paenibacillus macerans mutant N603D | | Descriptor: | CALCIUM ION, Cyclomaltodextrin glucanotransferase | | Authors: | Li, C.M, Ban, X.F, Li, Z.F, Li, Y.L, Cheng, S.D, Zhang, C.Y, Jin, T.C, Gu, Z.B. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Cyclodextrin glycosyltransferase from Paenibacillus macerans mutant N603D
To Be Published
|
|
8Z0H
 
 | | Crystal structure of 9-mer peptide from ALV-J in complex with BF2*0201 | | Descriptor: | Beta-2-microglobulin, Gag protein, MHC class I alpha chain 2 | | Authors: | Jia, Y.S, Ma, M.L, Li, Y.L, Liao, M, Dai, M.M. | | Deposit date: | 2024-04-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure and Mechanism of ALV-J Epitope Presented by MHC Class I Molecule BF20201
Xu Mu Shou Yi Xue Bao, 55, 2024
|
|
8CX0
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC monomeric complex | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
8CX2
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 2 | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
8CX1
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 1 | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
9JN4
 
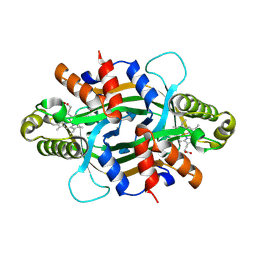 | | Crystal structure of KtzT-C197A in complex with HEME | | Descriptor: | FMN-binding protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, Y.L, Yang, Y.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-09-22 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Insights into the N-N Bond-Formation Mechanism of the Heme-Dependent Piperazate Synthase KtzT
Acs Catalysis, 15, 2025
|
|
9JN6
 
 | | Crystal structure of KtzT-C197A in complex with HEME and product | | Descriptor: | (3~{S})-1,2-diazinane-3-carboxylic acid, FMN-binding protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, Y.L, Yang, Y.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-09-22 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Insights into the N-N Bond-Formation Mechanism of the Heme-Dependent Piperazate Synthase KtzT
Acs Catalysis, 15, 2025
|
|
9JN5
 
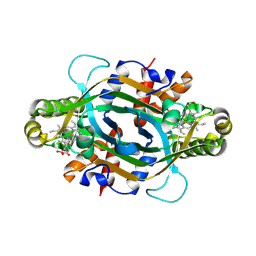 | | Crystal structure of KtzT-C197A in complex with HEME and substrate | | Descriptor: | FMN-binding protein, N~5~-hydroxy-L-ornithine, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, Y.L, Yang, Y.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-09-22 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the N-N Bond-Formation Mechanism of the Heme-Dependent Piperazate Synthase KtzT
Acs Catalysis, 15, 2025
|
|
8I6K
 
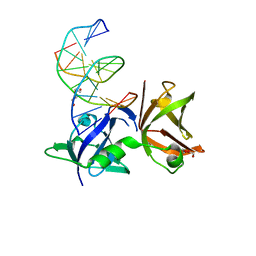 | | Structure of hMNDA HIN with dsDNA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Li, Y.L, Jin, T.C. | | Deposit date: | 2023-01-28 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism of dsDNA recognition by the hMNDA HIN domain: New insights into the DNA-binding model of a PYHIN protein.
Int.J.Biol.Macromol., 245, 2023
|
|
7KDT
 
 | |
9J8U
 
 | |
9J8T
 
 | |
1HJ1
 
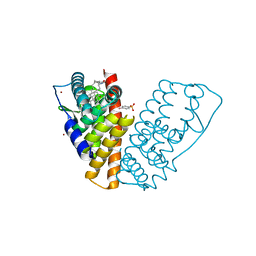 | | RAT OESTROGEN RECEPTOR BETA LIGAND-BINDING DOMAIN IN COMPLEX WITH PURE ANTIOESTROGEN ICI164,384 | | Descriptor: | N-BUTYL-11-[(7R,8R,9S,13S,14S,17S)-3,17-DIHYDROXY-13-METHYL-7,8,9,11,12,13,14,15,16,17-DECAHYDRO-6H-CYCLOPENTA[A]PHENANTHREN-7-YL]-N-METHYLUNDECANAMIDE, NICKEL (II) ION, OESTROGEN RECEPTOR BETA, ... | | Authors: | Pike, A.C.W, Brzozowski, A.M, Carlquist, M. | | Deposit date: | 2001-01-08 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights Into the Mode of Action of a Pure Antiestrogen
Structure, 9, 2001
|
|
6LKA
 
 | | Crystal Structure of EV71-3C protease with a Novel Macrocyclic Compounds | | Descriptor: | 3C proteinase, ~{N}-[(2~{S})-1-[[(2~{S},3~{S},6~{S},7~{Z},12~{E})-4,9-bis(oxidanylidene)-6-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]-2-phenyl-1,10-dioxa-5-azacyclopentadeca-7,12-dien-3-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Li, P, Wu, S.Q, Xiao, T.Y.C, Li, Y.L, Su, Z.M, Hao, F, Hu, G.P, Hu, J, Lin, F.S, Chen, X.S, Gu, Z.X, He, H.Y, Li, J, Chen, S.H. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Design, synthesis, and evaluation of a novel macrocyclic anti-EV71 agent.
Bioorg.Med.Chem., 28, 2020
|
|
7YEU
 
 | | Superfolder green fluorescent protein with phosphine unnatural amino acid P3BF | | Descriptor: | Superfolder green fluorescent protein | | Authors: | Hu, C, Duan, H.Z, Liu, X.H, Chen, Y.X, Wang, J.Y. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetically Encoded Phosphine Ligand for Metalloprotein Design.
J.Am.Chem.Soc., 144, 2022
|
|
7YK1
 
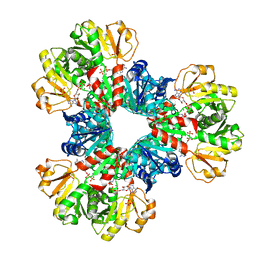 | | Structural basis of human PRPS2 filaments | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Lu, G.M, Hu, H.H, Liu, J.L. | | Deposit date: | 2022-07-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of human PRPS2 filaments.
Cell Biosci, 13, 2023
|
|
7VLN
 
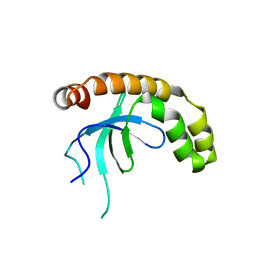 | | NSD2-PWWP1 domain bound with an imidazol-5-yl benzonitrile compound | | Descriptor: | 4-[5-[4-(aminomethyl)-2,6-dimethoxy-phenyl]-3-methyl-imidazol-4-yl]benzenecarbonitrile, Histone-lysine N-methyltransferase NSD2 | | Authors: | Cao, D.Y, Li, Y.L, Li, J, Xiong, B. | | Deposit date: | 2021-10-05 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Discovery of a Series of NSD2-PWWP1 Inhibitors.
J.Med.Chem., 65, 2022
|
|
