2N7J
 
 | |
7CNC
 
 | | cystal structure of human ERH in complex with DGCR8 | | 分子名称: | Enhancer of rudimentary homolog, Microprocessor complex subunit DGCR8 | | 著者 | Li, F, Shen, S. | | 登録日 | 2020-07-30 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | ERH facilitates microRNA maturation through the interaction with the N-terminus of DGCR8.
Nucleic Acids Res., 48, 2020
|
|
4GNE
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-7 | | 分子名称: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | 著者 | Li, F, He, C, Wu, J, Shi, Y. | | 登録日 | 2012-08-17 | | 公開日 | 2013-01-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GND
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains | | 分子名称: | Histone-lysine N-methyltransferase NSD3, ZINC ION | | 著者 | Li, F, He, C, Wu, J, Shi, Y. | | 登録日 | 2012-08-17 | | 公開日 | 2013-01-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GNF
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-15 | | 分子名称: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | 著者 | Li, F, He, C, Wu, J, Shi, Y. | | 登録日 | 2012-08-17 | | 公開日 | 2013-01-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GNG
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3K9me3 peptide | | 分子名称: | GLYCEROL, Histone H3.3, Histone-lysine N-methyltransferase NSD3, ... | | 著者 | Li, F, He, C, Wu, J, Shi, Y. | | 登録日 | 2012-08-17 | | 公開日 | 2013-01-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4H14
 
 | | Crystal Structure of Bovine Coronavirus Spike Protein Lectin Domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | 著者 | Li, F. | | 登録日 | 2012-09-10 | | 公開日 | 2012-10-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of bovine coronavirus spike protein lectin domain.
J.Biol.Chem., 287, 2012
|
|
2FIH
 
 | |
2FII
 
 | |
2FIJ
 
 | |
2FIL
 
 | |
5YJ3
 
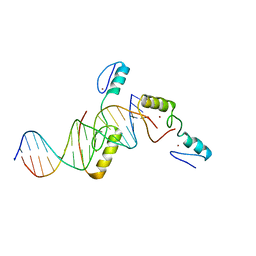 | | Crystal structure of TZAP and telomeric DNA complex | | 分子名称: | DNA (5'-D(*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), Telomere zinc finger-associated protein, ... | | 著者 | Li, F, Zhao, Y. | | 登録日 | 2017-10-07 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.845 Å) | | 主引用文献 | The 11th C2H2 zinc finger and an adjacent C-terminal arm are responsible for TZAP recognition of telomeric DNA.
Cell Res., 28, 2018
|
|
6AKJ
 
 | | The crystal structure of EMC complex | | 分子名称: | Enhancer of rudimentary homolog,YTH domain-containing protein mmi1 fusion protein, SULFATE ION | | 著者 | Li, F. | | 登録日 | 2018-09-01 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A conserved dimer interface connects ERH and YTH family proteins to promote gene silencing.
Nat Commun, 10, 2019
|
|
8J22
 
 | | Cryo-EM structure of FFAR2 complex bound with TUG-1375 | | 分子名称: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | 登録日 | 2023-04-14 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J21
 
 | | Cryo-EM structure of FFAR3 complex bound with butyrate acid | | 分子名称: | Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | 登録日 | 2023-04-14 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J20
 
 | | Cryo-EM structure of FFAR3 bound with valeric acid and AR420626 | | 分子名称: | (4R)-N-[2,5-bis(chloranyl)phenyl]-4-(furan-2-yl)-2-methyl-5-oxidanylidene-4,6,7,8-tetrahydro-1H-quinoline-3-carboxamide, Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | 登録日 | 2023-04-14 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J24
 
 | | Cryo-EM structure of FFAR2 complex bound with acetic acid | | 分子名称: | ACETATE ION, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Tai, L, Li, F, Tang, W, Sun, X, Wang, J. | | 登録日 | 2023-04-14 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
7WYG
 
 | | Crystal structure of P450BSbeta-L78I/Q85H/G290I variant in complex with palmitic acid. | | 分子名称: | Cytochrome P450 152A1, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | 著者 | Li, F, He, C, Wang, X. | | 登録日 | 2022-02-16 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Biocatalytic Enantioselective beta-Hydroxylation of Unactivated C-H Bonds in Aliphatic Carboxylic Acids.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8VQR
 
 | | Crystal structure of chimeric SARS-CoV-2 RBD complexed with chimeric raccoon dog ACE2 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hsueh, F.-C, Shi, K, Aihara, H, Li, F. | | 登録日 | 2024-01-19 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.565 Å) | | 主引用文献 | Structural basis for raccoon dog receptor recognition by SARS-CoV-2
To Be Published
|
|
7R1U
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the WZ16 inhibitor | | 分子名称: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | 登録日 | 2022-02-03 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
7R1T
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the SS148 inhibitor | | 分子名称: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | 登録日 | 2022-02-03 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
2QEG
 
 | | B-DNA with 7-deaza-dG modification | | 分子名称: | DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*DTP*DCP*(7GU)P*DCP*DG)-3') | | 著者 | Wang, F, Li, F, Ganguly, M, Marky, L.A, Gold, B, Egli, M, Stone, M.P. | | 登録日 | 2007-06-25 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A bridging water anchors the tethered 5-(3-aminopropyl)-2'-deoxyuridine amine in the DNA major groove proximate to the N+2 C.G base pair: implications for formation of interstrand 5'-GNC-3' cross-links by nitrogen mustards.
Biochemistry, 47, 2008
|
|
2QEF
 
 | | X-ray structure of 7-deaza-dG and Z3dU modified duplex CGCGAATXCZCG | | 分子名称: | DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*(ZDU)P*DCP*(7GU)P*DCP*DG)-3') | | 著者 | Wang, F, Li, F, Ganguly, M, Marky, L.A, Gold, B, Egli, M, Stone, M.P. | | 登録日 | 2007-06-25 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A bridging water anchors the tethered 5-(3-aminopropyl)-2'-deoxyuridine amine in the DNA major groove proximate to the N+2 C.G base pair: implications for formation of interstrand 5'-GNC-3' cross-links by nitrogen mustards.
Biochemistry, 47, 2008
|
|
4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | 分子名称: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | 著者 | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | 登録日 | 2014-11-04 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
4XKL
 
 | | Crystal structure of NDP52 ZF2 in complex with mono-ubiquitin | | 分子名称: | ACETATE ION, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | 著者 | Xie, X, Li, F, Wang, Y, Lin, Z, Chen, X, Liu, J, Pan, L. | | 登録日 | 2015-01-12 | | 公開日 | 2015-11-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2
Autophagy, 11, 2015
|
|
