2MX0
 
 | | Solution structure of HP0268 from Helicobacter pylori | | Descriptor: | Uncharacterized protein HP_0268 | | Authors: | Lee, K.Y. | | Deposit date: | 2014-12-05 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-based functional identification of Helicobacter pylori HP0268 as a nuclease with both DNA nicking and RNase activities
Nucleic Acids Res., 43, 2015
|
|
8YPJ
 
 | |
1KMR
 
 | |
6OS4
 
 | | Calmodulin in complex with farnesyl cysteine methyl ester | | Descriptor: | CALCIUM ION, Calmodulin-1, s-farnesyl-l-cysteine methyl ester | | Authors: | Grant, B.M.M, Enomoto, M, Lee, K.Y, Back, S.I, Gebregiworgis, T, Ishiyama, N, Ikura, M, Marshall, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Calmodulin disrupts plasma membrane localization of farnesylated KRAS4b by sequestering its lipid moiety.
Sci.Signal., 13, 2020
|
|
1DFW
 
 | | CONFORMATIONAL MAPPING OF THE N-TERMINAL SEGMENT OF SURFACTANT PROTEIN B IN LIPID USING 13C-ENHANCED FOURIER TRANSFORM INFRARED SPECTROSCOPY (FTIR) | | Descriptor: | LUNG SURFACTANT PROTEIN B | | Authors: | Gordon, L.M, Lee, K.Y.C, Lipp, M.M, Zasadzinski, J.A, Walther, F.J, Sherman, M.A, Waring, A.J. | | Deposit date: | 1999-11-22 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-07 | | Method: | INFRARED SPECTROSCOPY | | Cite: | Conformational mapping of the N-terminal segment of surfactant protein B in lipid using 13C-enhanced Fourier transform infrared spectroscopy.
J.Pept.Res., 55, 2000
|
|
6W4F
 
 | | NMR-driven structure of KRAS4B-GDP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6W4E
 
 | | NMR-driven structure of KRAS4B-GTP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7RSE
 
 | | NMR-driven structure of the KRAS4B-G12D "alpha-beta" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
7RSC
 
 | | NMR-driven structure of the KRAS4B-G12D "alpha-alpha" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
5XE2
 
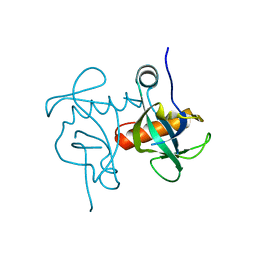 | | Endoribonuclease from Mycobacterial species | | Descriptor: | Endoribonuclease MazF4 | | Authors: | Ahn, D.-H, Lee, K.-Y, Lee, S.J, Yoon, H.J, Kim, S.-J, Lee, B.-J. | | Deposit date: | 2017-03-31 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural analyses of the MazEF4 toxin-antitoxin pair in Mycobacterium tuberculosis provide evidence for a unique extracellular death factor.
J. Biol. Chem., 292, 2017
|
|
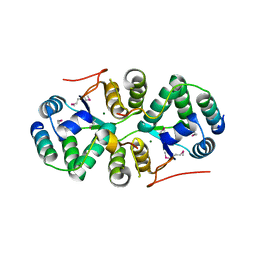
4XGQ
 
 | | Crystal structure of addiction module from Mycobacterial species | | Descriptor: | Antitoxin VapB30, MAGNESIUM ION, Ribonuclease VapC30 | | Authors: | Lee, B.-J, Lee, I.-G, Lee, S.J. | | Deposit date: | 2015-01-02 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional studies of the Mycobacterium tuberculosis VapBC30 toxin-antitoxin system: implications for the design of novel antimicrobial peptides
Nucleic Acids Res., 43, 2015
|
|
4XGR
 
 | | Crystal structure of addiction module from Mycobacterial species | | Descriptor: | Antitoxin VapB30, MAGNESIUM ION, Ribonuclease VapC30 | | Authors: | Lee, B.-J, Lee, I.-G, Lee, S.J. | | Deposit date: | 2015-01-02 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional studies of the Mycobacterium tuberculosis VapBC30 toxin-antitoxin system: implications for the design of novel antimicrobial peptides
Nucleic Acids Res., 43, 2015
|
|
5X3T
 
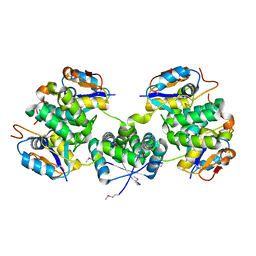 | | VapBC from Mycobacterium tuberculosis | | Descriptor: | Antitoxin VapB26, MAGNESIUM ION, Ribonuclease VapC26 | | Authors: | Kang, S.M, Kim, D.H, Yoon, H.J, Lee, B.J. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional details of the Mycobacterium tuberculosis VapBC26 toxin-antitoxin system based on a structural study: insights into unique binding and antibiotic peptides.
Nucleic Acids Res., 45, 2017
|
|
5XR2
 
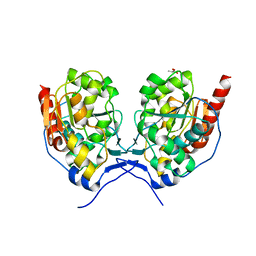 | | SAV0551 | | Descriptor: | LACTIC ACID, Protein/nucleic acid deglycase HchA, ZINC ION | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2017-06-07 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional studies of SAV0551 fromStaphylococcus aureusas a chaperone and glyoxalase III.
Biosci. Rep., 37, 2017
|
|
4QXZ
 
 | |
3UI3
 
 | |
6JKG
 
 | | The NAD+-free form of human NSDHL | | Descriptor: | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
6JKH
 
 | | The NAD+-bound form of human NSDHL | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
4Z9E
 
 | | Alba from Thermoplasma volcanium | | Descriptor: | DNA/RNA-binding protein Alba | | Authors: | Ma, C, Lee, S.J, Pathak, C, Lee, B.J. | | Deposit date: | 2015-04-10 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Alba from Thermoplasma volcanium belongs to alpha-NAT's: An insight into the structural aspects of Tv Alba and its acetylation by Tv Ard1.
Arch.Biochem.Biophys., 590, 2016
|
|
4Z1B
 
 | | Structure of H204A mutant KDO8PS from H.pylori | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase | | Authors: | Lee, B.J, Cho, S, Im, H, Yoon, H.J. | | Deposit date: | 2015-03-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
Eur.J.Med.Chem., 108, 2016
|
|
4Z1C
 
 | | Structure of Cadmium bound KDO8PS from H.pylori | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION | | Authors: | Lee, B.J, Cho, S, Im, H, Yoon, H.J. | | Deposit date: | 2015-03-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
Eur.J.Med.Chem., 108, 2016
|
|
4Z1A
 
 | | Structure of apo form KDO8PS from H.pylori | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase | | Authors: | Lee, B.J, Cho, S, Im, H, Yoon, H.J. | | Deposit date: | 2015-03-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
Eur.J.Med.Chem., 108, 2016
|
|
4Z1D
 
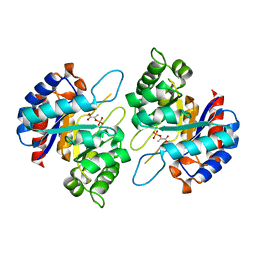 | | Structure of PEP and zinc bound KDO8PS from H.pylori | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, PHOSPHOENOLPYRUVATE, ZINC ION | | Authors: | Lee, B.J, Cho, S, Im, H, Yoon, H.J. | | Deposit date: | 2015-03-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
Eur.J.Med.Chem., 108, 2016
|
|
6PTS
 
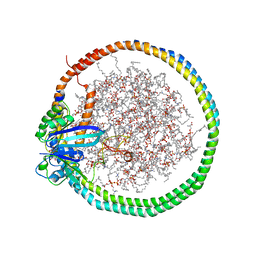 | | NMR data-driven model of KRas-GMPPNP:RBD-CRD complex tethered to a nanodisc (state A) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Lee, K, Gasmi-Seabrook, G, Ikura, M, Marshall, C.B. | | Deposit date: | 2019-07-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PTW
 
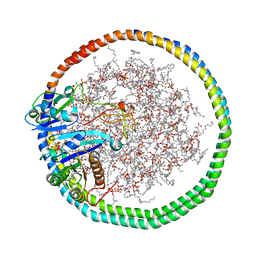 | | NMR data-driven model of KRas-GMPPNP:RBD-CRD complex tethered to a nanodisc (state B) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Lee, K, Gasmi-Seabrook, G, Ikura, M, Marshall, C.B. | | Deposit date: | 2019-07-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
