7F5Y
 
 | | Crystal structure of the single-stranded dna-binding protein from Mycobacterium tuberculosis- Form III | | Descriptor: | FORMIC ACID, Single-stranded DNA-binding protein | | Authors: | Srikalaivani, R, Paul, A, Sriram, R, Narayanan, S, Gopal, B, Vijayan, M. | | Deposit date: | 2021-06-23 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural variability of Mycobacterium tuberculosis SSB and susceptibility to inhibition.
Curr.Sci., 122, 2022
|
|
7F5Z
 
 | | Crystal structure of the single-stranded dna-binding protein from Mycobacterium tuberculosis- Form III | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Srikalaivani, R, Paul, A, Sriram, R, Narayanan, S, Gopal, B, Vijayan, M. | | Deposit date: | 2021-06-23 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural variability of Mycobacterium tuberculosis SSB and susceptibility to inhibition.
Curr.Sci., 122, 2022
|
|
5Y8O
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + NAD + 3-Hydroxy propionate (3-HP) | | Descriptor: | (2~{S})-2-methylpentanedioic acid, 3-HYDROXY-PROPANOIC ACID, ACRYLIC ACID, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8I
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + (S)-3-hydroxyisobutyrate (S-HIBA) | | Descriptor: | (2S)-2-methyl-3-oxidanyl-propanoic acid, (2~{S})-2-methylpentanedioic acid, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8P
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + 3-Hydroxy propionate (3-HP) | | Descriptor: | (2~{S})-2-methylpentanedioic acid, 3-HYDROXY-PROPANOIC ACID, ACRYLIC ACID, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8K
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + L-serine | | Descriptor: | (2~{S})-2-methylpentanedioic acid, ACRYLIC ACID, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8M
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + NAD + (R)-3-hydroxyisobutyrate (R-HIBA) | | Descriptor: | (2R)-3-HYDROXY-2-METHYLPROPANOIC ACID, (2~{S})-2-methylpentanedioic acid, ACRYLIC ACID, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8G
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate Dehydrogenase (MtHIBADH) | | Descriptor: | (2~{S})-2-methylpentanedioic acid, ACRYLIC ACID, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8J
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + (R)-3-hydroxyisobutyrate (R-HIBA) | | Descriptor: | (2R)-3-HYDROXY-2-METHYLPROPANOIC ACID, (2~{S})-2-methylpentanedioic acid, ACRYLIC ACID, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8L
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + NAD +(S)-3-hydroxyisobutyrate (S-HIBA) | | Descriptor: | (2S)-2-methyl-3-oxidanyl-propanoic acid, (2~{S})-2-methylpentanedioic acid, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8N
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + NAD + L-serine | | Descriptor: | (2~{S})-2-methylpentanedioic acid, ACRYLIC ACID, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8H
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + NAD+ | | Descriptor: | (2~{S})-2-methylpentanedioic acid, ACRYLIC ACID, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
1YJE
 
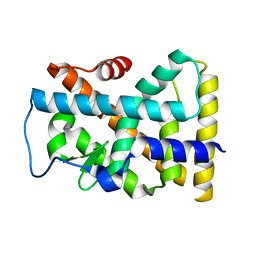 | | Crystal structure of the rNGFI-B ligand-binding domain | | Descriptor: | Orphan nuclear receptor NR4A1 | | Authors: | Flaig, R, Greschik, H, Peluso-Iltis, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain.
J.Biol.Chem., 280, 2005
|
|
5X0S
 
 | | Solution NMR structure of peptide toxin SsTx from Scolopendra subspinipes mutilans | | Descriptor: | SsTx | | Authors: | Wu, F, Luo, L, Qu, D, Zhang, L, Tian, C, Lai, R. | | Deposit date: | 2017-01-23 | | Release date: | 2018-01-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Centipedes subdue giant prey by blocking KCNQ channels
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6LF5
 
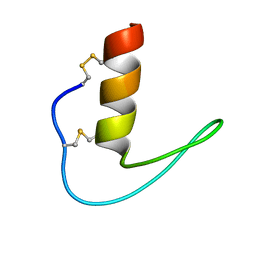 | | The solution structure of ShSPI | | Descriptor: | ShSPI | | Authors: | Luan, N, Rong, M.Q, Liu, J.X, Lai, R. | | Deposit date: | 2019-11-29 | | Release date: | 2020-12-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and Characterization of ShSPI, a Kazal-Type Elastase Inhibitor from the Venom of Scolopendra Hainanum .
Toxins, 11, 2019
|
|
2O9Q
 
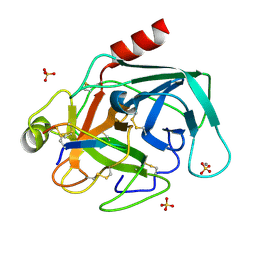 | | The crystal structure of Bovine Trypsin complexed with a small inhibition peptide ORB2K | | Descriptor: | CALCIUM ION, Cationic trypsin, ORB2K, ... | | Authors: | Li, J, Zhang, C, Xu, X, Wang, J, Gong, W, Lai, R. | | Deposit date: | 2006-12-14 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | From protease inhibitor to antibiotics: single point mutation makes tremendous functional shift
To be Published
|
|
2M35
 
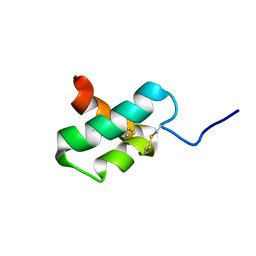 | | NMR study of k-Ssm1a | | Descriptor: | k-Ssm1a | | Authors: | King, G.F, Undheim, E.A, Mobli, M, Yang, S, Rong, M, Lai, R. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR study of k-Ssm1a
To be Published
|
|
2JQW
 
 | |
2KCR
 
 | |
1TFC
 
 | | CRYSTAL STRUCTURE OF THE LIGAND-BINDING DOMAIN OF THE ESTROGEN-RELATED RECEPTOR GAMMA IN COMPLEX WITH A STEROID RECEPTOR COACTIVATOR-1 PEPTIDE | | Descriptor: | Estrogen-related receptor gamma, steroid receptor coactivator-1 | | Authors: | Greschik, H, Flaig, R, Renaud, J.P, Moras, D. | | Deposit date: | 2004-05-27 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Deactivation of the Estrogen-related Receptor {gamma} by Diethylstilbestrol or 4-Hydroxytamoxifen and Determinants of Selectivity.
J.Biol.Chem., 279, 2004
|
|
5J6S
 
 | | Crystal structure of Endoplasmic Reticulum Aminopeptidase 2 (ERAP2) in complex with a hydroxamic derivative ligand | | Descriptor: | (2S)-N~1~-benzyl-2-[(4-fluorophenyl)methyl]-N~3~-hydroxypropanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saridakis, E, Giastas, P, Mpakali, A, Deprez-Poulain, R, Stratikos, E. | | Deposit date: | 2016-04-05 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency.
ACS Med Chem Lett, 8, 2017
|
|
2YPU
 
 | | human insulin degrading enzyme E111Q in complex with inhibitor compound 41367 | | Descriptor: | 2-[[2-[[(2S)-3-(3H-IMIDAZOL-4-YL)-1-METHOXY-1-OXO-PROPAN-2-YL]AMINO]-2-OXO-ETHYL]-(PHENYLMETHYL)AMINO]ETHANOIC ACID, INSULIN-DEGRADING ENZYME, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.-J. | | Deposit date: | 2012-11-01 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Imidazole-Derived 2-[N-Carbamoylmethyl-Alkylamino]Acetic Acids,Substrate-Dependent Modulators of Insulin-Degrading Enzyme in Amyloid-Beta Hydrolysis
Eur J Med Chem, 79C, 2014
|
|
5AFW
 
 | | Assembly of methylated LSD1 and CHD1 drives AR-dependent transcription and translocation | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 1, ... | | Authors: | Metzger, E, Willmann, D, McMillan, J, Petroll, K, Metzger, P, Gerhardt, S, vonMaessenhausen, A, Schott, A.K, Espejo, A, Eberlin, A, Wohlwend, D, Schuele, K.M, Schleicher, M, Perner, S, Bedford, M.T, Dengjel, J, Flaig, R, Einsle, O, Schuele, R. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Assembly of Methylated Kdm1A and Chd1 Drives Androgen Receptor-Dependent Transcription and Translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3G1M
 
 | | EthR from Mycobacterium tuberculosis in complex with compound BDM31381 | | Descriptor: | 1-(thiophen-2-ylacetyl)-4-(3-thiophen-2-yl-1,2,4-oxadiazol-5-yl)piperidine, TRANSCRIPTIONAL REGULATORY REPRESSOR PROTEIN (TETR-FAMILY) ETHR | | Authors: | Willand, N, Dirie, B, Carette, X, Bifani, P, Singhal, A, Desroses, M, Leroux, F, Willery, E, Mathys, V, Deprez-Poulain, R, Delcroix, G, Frenois, F, Aumercier, M, Locht, C, Villeret, V, Deprez, B, Baulard, A.R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthetic EthR inhibitors boost antituberculous activity of ethionamide.
Nat.Med. (N.Y.), 15, 2009
|
|
3G1O
 
 | | EthR from Mycobacterium tuberculosis in complex with compound BDM14500 | | Descriptor: | TRANSCRIPTIONAL REGULATORY REPRESSOR PROTEIN (TETR-FAMILY) ETHR, tert-butyl 4-(3-thiophen-2-yl-1,2,4-oxadiazol-5-yl)piperidine-1-carboxylate | | Authors: | Willand, N, Dirie, B, Carette, X, Bifani, P, Singhal, A, Desroses, M, Leroux, F, Willery, E, Mathys, V, Deprez-Poulain, R, Delcroix, G, Frenois, F, Aumercier, M, Locht, C, Villeret, V, Deprez, B, Baulard, A.R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthetic EthR inhibitors boost antituberculous activity of ethionamide.
NAT.MED. (N.Y.), 15, 2009
|
|
