3EKA
 
 | | Crystal structure of the complex of hyaluranidase trimer with ascorbic acid at 3.1 A resolution reveals the locations of three binding sites | | Descriptor: | ASCORBIC ACID, Hyaluronidase, phage associated | | Authors: | Mishra, P, Ethayathulla, A.S, Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Bhakuni, V, Singh, T.P. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Polysaccharide binding sites in hyaluronate lyase--crystal structures of native phage-encoded hyaluronate lyase and its complexes with ascorbic acid and lactose.
Febs J., 276, 2009
|
|
3FO7
 
 | | Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site | | Descriptor: | INDOMETHACIN, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Prem Kumar, R, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site
To be Published
|
|
6I1T
 
 | | Calcium structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Turbe-Doan, A, Record, E, Lombard, V, Kumar, R, Henrissat, B, Levasseur, A, Garron, M.L. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The functional and structural characterization ofTrichoderma reeseidehydrogenase belonging to the PQQ dependent family of Carbohydrate-Active Enzymes Family AA12.
Appl.Environ.Microbiol., 2019
|
|
6I1Q
 
 | | Iodide structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Turbe-Doan, A, Record, E, Lombard, V, Kumar, R, Henrissat, B, Levasseur, A, Garron, M.L. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The functional and structural characterization ofTrichoderma reeseidehydrogenase belonging to the PQQ dependent family of Carbohydrate-Active Enzymes Family AA12.
Appl.Environ.Microbiol., 2019
|
|
4FAO
 
 | | Specificity and Structure of a high affinity Activin-like 1 (ALK1) signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 2, ... | | Authors: | Townson, S.A, Martinez-Hackert, E, Greppi, C, Lowden, P, Sako, D, Liu, J, Ucran, J.A, Liharska, K, Underwood, K.W, Seehra, J, Kumar, R, Grinberg, A.V. | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.357 Å) | | Cite: | Specificity and Structure of a High Affinity Activin Receptor-like Kinase 1 (ALK1) Signaling Complex.
J.Biol.Chem., 287, 2012
|
|
1ZWP
 
 | | The atomic resolution Crystal structure of the Phospholipase A2 (PLA2) complex with Nimesulide reveals its weaker binding to PLA2 | | Descriptor: | 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2005-06-04 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The atomic resolution Crystal structure of the Phospholipase A2 (PLA2) complex with Nimesulide reveals its weaker binding to PLA2
To be Published
|
|
2B65
 
 | | Crystal structure of the complex of C-lobe of bovine lactoferrin with maltose at 1.5A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Singh, N, Prem kumar, R, Jabeen, T, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the complex of C-lobe of bovine lactoferrin with maltose at 1.5A resolution
To be published
|
|
2PGH
 
 | | STRUCTURE DETERMINATION OF AQUOMET PORCINE HEMOGLOBIN AT 2.8 ANGSTROM RESOLUTION | | Descriptor: | HEMOGLOBIN (AQUO MET) (ALPHA CHAIN), HEMOGLOBIN (AQUO MET) (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Katz, D.S, White, S.P, Huang, W, Kumar, R, Christianson, D.W. | | Deposit date: | 1994-09-16 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure determination of aquomet porcine hemoglobin at 2.8 A resolution.
J.Mol.Biol., 244, 1994
|
|
2DP5
 
 | | Structure of streptococcus pyogenes bacteriophage-associated hyaluronate lyase Hylp2 | | Descriptor: | Hyaluronidase | | Authors: | Mishra, P, Bhakuni, V, Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2006-05-06 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure of streptococcus pyogenes bacteriophage-associated hyaluronate lyase Hylp2
To be Published
|
|
2EFB
 
 | | Crystal structure of the complex of goat lactoperoxidase with phosphate at 2.94 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Hariprasad, G, Prem Kumar, R, Singh, N, Bhushan, A, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with phosphate at 2.94 A resolution
To be Published
|
|
2E9E
 
 | | Crystal structure of the complex of goat lactoperoxidase with Nitrate at 3.25 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Prem kumar, R, Singh, N, Sharma, S, Singh, S.B, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with Nitrate at 3.25 A resolution
To be Published
|
|
7LXD
 
 | | Structure of yeast DNA Polymerase Zeta (apo) | | Descriptor: | DNA polymerase delta small subunit, DNA polymerase delta subunit 3, DNA polymerase zeta catalytic subunit, ... | | Authors: | Truong, C.D, Craig, T.A, Cui, G, Botuyan, M.V, Serkasevich, R.A, Chan, K.-Y, Mer, G, Chiu, P.-L, Kumar, R. | | Deposit date: | 2021-03-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Cryo-EM reveals conformational flexibility in apo DNA polymerase zeta.
J.Biol.Chem., 297, 2021
|
|
4XMK
 
 | |
4XML
 
 | | Crystal structure of Fab of HIV-1 gp120 V3-specific human monoclonal antibody 2424 | | Descriptor: | Heavy chain of HIV-1 gp120 V3-specific human monoclonal antibody 2424, Light chain of HIV-1 gp120 V3-specific human monoclonal antibody 2424 | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2015-01-14 | | Release date: | 2015-07-08 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Functional and Structural Characterization of Human V3-Specific Monoclonal Antibody 2424 with Neutralizing Activity against HIV-1 JRFL.
J.Virol., 89, 2015
|
|
4E8G
 
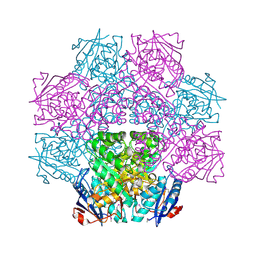 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound mg | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, N-terminal domain protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
6E1J
 
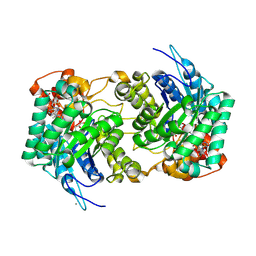 | |
4IZG
 
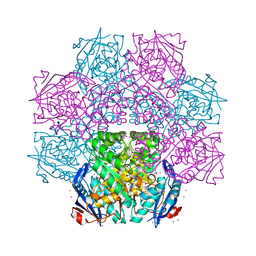 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound cis-4oh-d-proline betaine (product) | | Descriptor: | (2S,4S)-2-carboxy-4-hydroxy-1,1-dimethylpyrrolidinium, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, LaFleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
4J1O
 
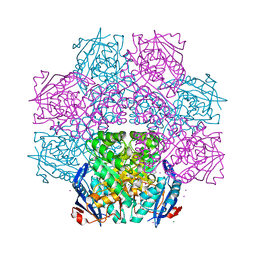 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound l-proline betaine (substrate) | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, GLYCEROL, IODIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, LaFleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
6HFG
 
 | |
8ORC
 
 | | Mus Musculus Acetylcholinesterase in complex with AL237 | | Descriptor: | 1-[2-(dimethylamino)ethyl]-3-(2-methoxyphenyl)thiourea, 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Ekstrom, F.E, Linusson, A. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzyme Dynamics Determine Potency and Selectivity of Inhibitors Targeting Disease-Transmitting Mosquitoes
To Be Published
|
|
1B99
 
 | | 3'-FLUORO-URIDINE DIPHOSPHATE BINDING TO NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | 2',3'-DIDEOXY-3'-FLUORO-URIDIDINE-5'-DIPHOSPHATE, PROTEIN (NUCLEOSIDE DIPHOSPHATE KINASE), PYROPHOSPHATE 2- | | Authors: | Janin, J, Xu, Y. | | Deposit date: | 1999-02-22 | | Release date: | 1999-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of nucleoside diphosphate kinase investigated using nucleotide analogues, viscosity effects, and X-ray crystallography.
Biochemistry, 38, 1999
|
|
5OJL
 
 | | Imine Reductase from Aspergillus terreus in complex with NADPH4 and dibenz[c,e]azepine | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, 5-methyl-7~{H}-benzo[d][2]benzazepine, Imine reductase | | Authors: | Sharma, M, Grogan, G. | | Deposit date: | 2017-07-21 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Biocatalytic Routes to Enantiomerically Enriched Dibenz[c,e]azepines.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3KJZ
 
 | | Crystal structure of native peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, N, Yadav, R, Prem Kumar, R, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2009-11-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from mycobacterium smegmatis reveals novel features related to enzyme dynamics.
Int J Biochem Mol Biol, 3, 2012
|
|
5JPV
 
 | |
7WVL
 
 | |
