8WC5
 
 | | Cryo-EM structure of the TMA-bound mTAAR1-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCA
 
 | | Cryo-EM structure of the PEA-bound hTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC3
 
 | | Cryo-EM structure of the SEP363856-bound mTAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC4
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC6
 
 | | Cryo-EM structure of the PEA-bound mTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCB
 
 | | Cryo-EM structure of the CHA-bound mTAAR1-Gq complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
6DUG
 
 | | Crystal structure of HIV-1 reverse transcriptase K101P mutant in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-06-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6DUH
 
 | | Crystal structure of HIV-1 reverse transcriptase Y181I mutant in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-06-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6DUF
 
 | | Crystal structure of HIV-1 reverse transcriptase V106A/F227L mutant in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-06-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
7QHE
 
 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((naphthalen-1-yl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | Descriptor: | (3~{S})-1-[[4-(naphthalen-1-ylcarbamoyl)phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
7QHD
 
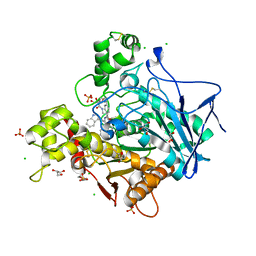 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((2-(1H-indol-3-yl)ethyl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | Descriptor: | (3~{S})-1-[[4-[2-(1~{H}-indol-3-yl)ethylcarbamoyl]phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
8FE8
 
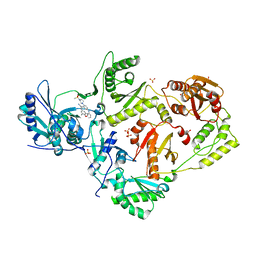 | | Crystal Structure of HIV-1 RT in Complex with the non-nucleoside inhibitor 18b1 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}pyrimidin-2-yl)amino]-2-[4-(methanesulfonyl)piperazin-1-yl]benzonitrile, Reverse transcriptase p51, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of diarylpyrimidine derivatives bearing piperazine sulfonyl as potent HIV-1 nonnucleoside reverse transcriptase inhibitors.
Commun Chem, 6, 2023
|
|
8JGF
 
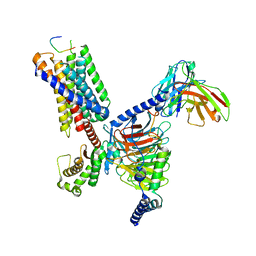 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide agonist BAM8-22 | | Descriptor: | BAM8-22, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Xu, H.E, Yang, F, Liu, Z.M, Guo, L.L, Zhang, Y.M, Fang, G.X, Tie, L, Zhuang, Y.M, Xue, C.Y. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ligand recognition and G protein coupling of the human itch receptor MRGPRX1.
Nat Commun, 14, 2023
|
|
8JGB
 
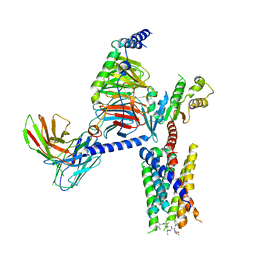 | | CryoEM structure of Gi-coupled MRGPRX1 with peptide agonist CNF-Tx2 | | Descriptor: | Conorfamide-Tx2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Xu, H.E, Yang, F, Liu, Z.M, Guo, L.L, Zhang, Y.M, Fang, G.X, Tie, L, Zhuang, Y.M, Xue, C.Y. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Ligand recognition and G protein coupling of the human itch receptor MRGPRX1.
Nat Commun, 14, 2023
|
|
8JGG
 
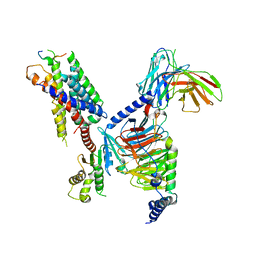 | | CryoEM structure of Gi-coupled MRGPRX1 with peptide agonist BAM8-22 | | Descriptor: | BAM8-22, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Xu, H.E, Ynag, F, Liu, Z.M, Guo, L.L, Zhang, Y.M, Fang, G.X, Tie, L, Zhuang, Y.M, Xue, C.Y. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand recognition and G protein coupling of the human itch receptor MRGPRX1.
Nat Commun, 14, 2023
|
|
6LIN
 
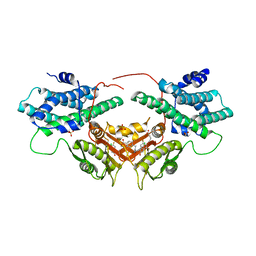 | | Crystal structure of human PDK2 complexed with GM10030 | | Descriptor: | 4-[[[4-[3,5-bis(fluoranyl)-4-(4-oxidanyl-4-oxidanylidene-butoxy)phenyl]-5-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-1,2-oxazol-3-yl]carbonylamino]methyl]benzoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kang, J, Kim, J. | | Deposit date: | 2019-12-12 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis for the inhibition of PDK2 by novel ATP- and lipoyl-binding site targeting compounds.
Biochem.Biophys.Res.Commun., 527, 2020
|
|
6LIO
 
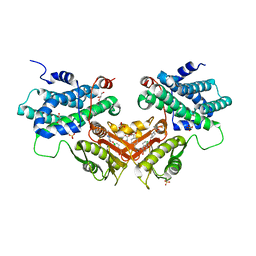 | | Crystal structure of human PDK2 complexed with GM67520 | | Descriptor: | 4-[[[5-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-[4-(1-methylsulfonylpiperidin-4-yl)oxyphenyl]-1,2-oxazol-3-yl]carbonylamino]methyl]cyclohexane-1-carboxylic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Kang, J, Kim, J. | | Deposit date: | 2019-12-12 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for the inhibition of PDK2 by novel ATP- and lipoyl-binding site targeting compounds.
Biochem.Biophys.Res.Commun., 527, 2020
|
|
6LIL
 
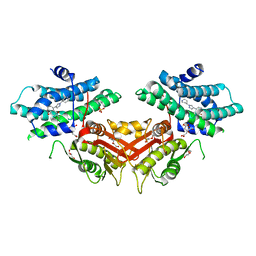 | |
2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | Descriptor: | Protein tyrosine phosphatase type IVA 3 | | Authors: | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
8B56
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GD-9 | | Descriptor: | (2~{S})-4-(2-chloranylethanoyl)-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5, BROMIDE ION, ... | | Authors: | Straeter, N, Muller, C.E, Claff, T, Sylvester, K, Weisse, R, Gao, S, Song, L, Liu, X, Zhan, P. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Discovery and Crystallographic Studies of Nonpeptidic Piperazine Derivatives as Covalent SARS-CoV-2 Main Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
3WW6
 
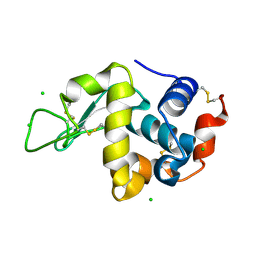 | | Crystal Structure of hen egg white lysozyme mutant N46D/D52S | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Abe, Y, Kubota, M, Ito, Y, Imoto, T, Ueda, T. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Effect on catalysis by replacement of catalytic residue from hen egg white lysozyme to Venerupis philippinarum lysozyme.
Protein Sci., 25, 2016
|
|
3WW5
 
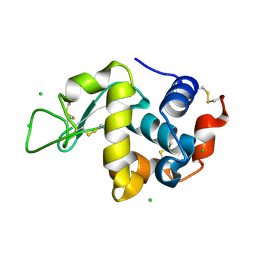 | | Crystal Structure of hen egg white lysozyme mutant N46E/D52S | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Abe, Y, Kubota, M, Ito, Y, Imoto, T, Ueda, T. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Effect on catalysis by replacement of catalytic residue from hen egg white lysozyme to Venerupis philippinarum lysozyme.
Protein Sci., 25, 2016
|
|
2MUN
 
 | |
8ACD
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GA-17S | | Descriptor: | (2~{S})-4-[[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-6-yl]carbonyl]-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
8ACL
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GC-14 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-pyridin-3-ylcarbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
