1FZV
 
 | | THE CRYSTAL STRUCTURE OF HUMAN PLACENTA GROWTH FACTOR-1 (PLGF-1), AN ANGIOGENIC PROTEIN AT 2.0A RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PLACENTA GROWTH FACTOR | | Authors: | Iyer, S, Leonidas, D.D, Swaminathan, G.J, Maglione, D, Battisti, M, Tucci, M, Persico, M.G, Acharya, K.R. | | Deposit date: | 2000-10-04 | | Release date: | 2001-05-09 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human placenta growth factor-1 (PlGF-1), an angiogenic protein, at 2.0 A resolution.
J.Biol.Chem., 276, 2001
|
|
2J0T
 
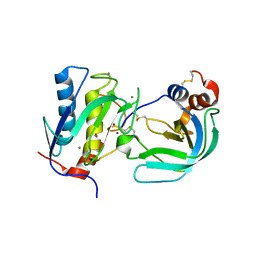 | | Crystal Structure of the Catalytic Domain of MMP-1 in Complex with the Inhibitory Domain of TIMP-1 | | Descriptor: | CALCIUM ION, INTERSTITIAL COLLAGENASE, METALLOPROTEINASE INHIBITOR 1, ... | | Authors: | Iyer, S, Wei, S, Brew, K, Acharya, K.R. | | Deposit date: | 2006-08-04 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Matrix Metalloproteinase-1 in Complex with the Inhibitory Domain of Tissue Inhibitor of Metalloproteinase-1.
J.Biol.Chem., 282, 2007
|
|
2XAC
 
 | |
4S2R
 
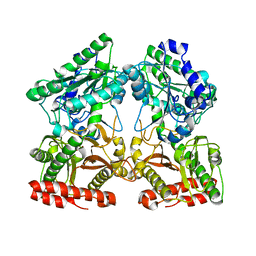 | | Crystal structure of X-prolyl aminopeptidase from Caenorhabditis elegans: a cytosolic enzyme with a di-nuclear active site | | Descriptor: | Protein APP-1, ZINC ION | | Authors: | Iyer, S, La-Borde, P, Payne, K.A.P, Parsons, M.R, Turner, A.J, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2015-01-22 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Crystal structure of X-prolyl aminopeptidase from Caenorhabditis elegans: A cytosolic enzyme with a di-nuclear active site.
FEBS Open Bio, 5, 2015
|
|
4S2T
 
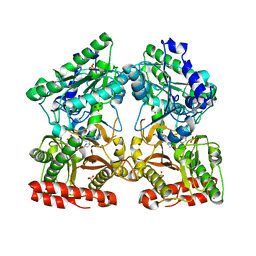 | | Crystal structure of X-prolyl aminopeptidase from Caenorhabditis elegans: a cytosolic enzyme with a di-nuclear active site | | Descriptor: | Protein APP-1, SULFATE ION, ZINC ION, ... | | Authors: | Iyer, S, La-Borde, P, Payne, K.A.P, Parsons, M.R, Turner, A.J, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2015-01-22 | | Release date: | 2015-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of X-prolyl aminopeptidase from Caenorhabditis elegans: A cytosolic enzyme with a di-nuclear active site.
FEBS Open Bio, 5, 2015
|
|
3ZBW
 
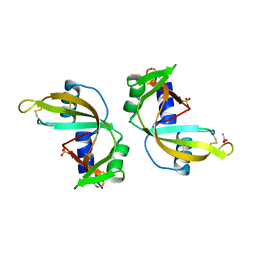 | | Crystal Structure of murine Angiogenin-3 | | Descriptor: | ACETIC ACID, ANGIOGENIN-3, SULFATE ION, ... | | Authors: | Iyer, S, Holloway, D.E, Acharya, K.R. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structures of Murine Angiogenin-2 and -3 - Probing 'Structure - Function' Relationships Amongst Angiogenin Homologues.
FEBS J., 280, 2013
|
|
3ZBV
 
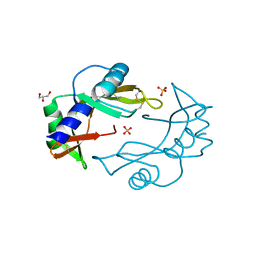 | | Crystal Structure of murine Angiogenin-2 | | Descriptor: | ANGIOGENIN-2, GLYCEROL, SULFATE ION, ... | | Authors: | Iyer, S, Holloway, D.E, Acharya, K.R. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of Murine Angiogenin-2 and -3 - Probing 'Structure - Function' Relationships Amongst Angiogenin Homologues.
FEBS J., 280, 2013
|
|
2BEX
 
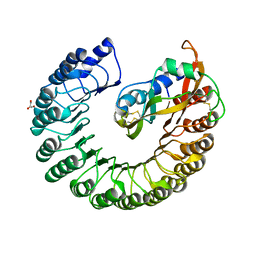 | | Crystal structure of Placental Ribonuclease Inhibitor in complex with Human Eosinophil Derived Neurotoxin at 2A resolution | | Descriptor: | ALPHA-KETOMALONIC ACID, GLYCEROL, NONSECRETORY RIBONUCLEASE, ... | | Authors: | Iyer, S, Holloway, D.E, Kumar, K, Shapiro, R, Acharya, K.R. | | Deposit date: | 2004-12-01 | | Release date: | 2005-03-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular Recognition of Human Eosinophil-Derived Neurotoxin (Rnase 2) by Placental Ribonuclease Inhibitor
J.Mol.Biol., 347, 2005
|
|
2C7W
 
 | | Crystal Structure of human vascular endothelial growth factor-B: Identification of amino acids important for angiogeninc activity | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, VASCULAR ENDOTHELIAL GROWTH FACTOR B PRECURSOR | | Authors: | Iyer, S, Scotney, P.D, Nash, A.D, Acharya, K.R. | | Deposit date: | 2005-11-29 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of Human Vascular Endothelial Growth Factor-B: Identification of Amino Acids Important for Receptor Binding
J.Mol.Biol., 359, 2006
|
|
2CLT
 
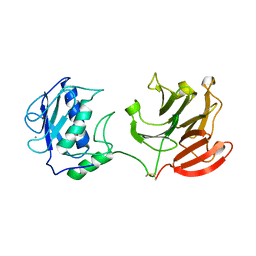 | | Crystal structure of the active form (full-length) of human fibroblast collagenase. | | Descriptor: | CALCIUM ION, INTERSTITIAL COLLAGENASE, ZINC ION | | Authors: | Iyer, S, Visse, R, Nagase, H, Acharya, K.R. | | Deposit date: | 2006-04-29 | | Release date: | 2006-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal Structure of an Active Form of Human Mmp-1.
J.Mol.Biol., 362, 2006
|
|
8TXO
 
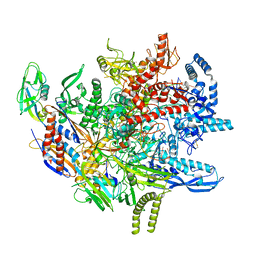 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound to the unnatural dZ-PTP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Aiyer, S, Shan, Z, Oh, J, Wang, D, Tan, Y.Z, Lyumkis, D. | | Deposit date: | 2023-08-23 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Overcoming resolution attenuation during tilted cryo-EM data collection.
Nat Commun, 15, 2024
|
|
7JYN
 
 | | Solution NMR structure of human Brd3 ET complexed with NSD3(148-184) peptide | | Descriptor: | Bromodomain-containing protein 3, Histone-lysine N-methyltransferase NSD3 | | Authors: | Aiyer, S, Swapna, G.V.T, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
7JQ8
 
 | | Solution NMR structure of human Brd3 ET domain | | Descriptor: | Bromodomain-containing protein 3, Integrase peptide | | Authors: | Aiyer, S, Swapna, G.V.T, Roth, J.M, Montelione, G.T. | | Deposit date: | 2020-08-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
7JMY
 
 | |
7JYZ
 
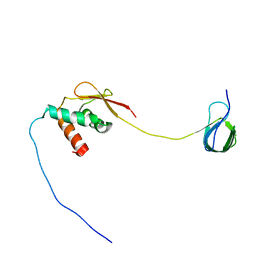 | | Solution NMR structure and dynamics of human Brd3 ET in complex with MLV IN CTD | | Descriptor: | Bromodomain-containing protein 3, Integrase | | Authors: | Aiyer, S, Liu, G, Swapna, G.V.T, Hao, J, Ma, L.C, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-09-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
2M9U
 
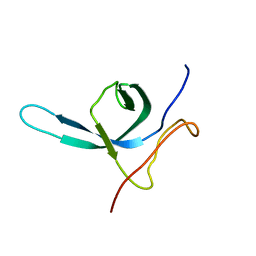 | | Solution NMR structure of the C-terminal domain (CTD) of Moloney murine leukemia virus integrase, Northeast Structural Genomics Target OR41A | | Descriptor: | Integrase p46 | | Authors: | Aiyer, S, Rossi, P, Schneider, W.M, Chander, A, Roth, M.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Altering murine leukemia virus integration through disruption of the integrase and BET protein family interaction.
Nucleic Acids Res., 42, 2014
|
|
6ULH
 
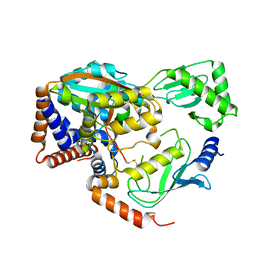 | |
6UMS
 
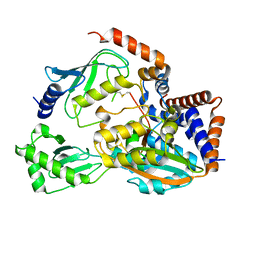 | |
6P5H
 
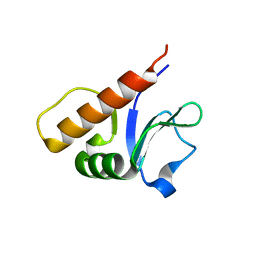 | |
6P5B
 
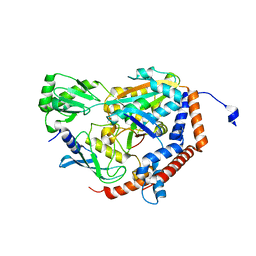 | | Crystal Structure of MavC in Complex with Ub-UbE2N | | Descriptor: | MavC, Ubiquitin, Ubiquitin-conjugating enzyme E2 N | | Authors: | Puvar, K, Iyer, S, Negron Teron, K.I, Das, C. | | Deposit date: | 2019-05-30 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Legionella effector MavC targets the Ube2N~Ub conjugate for noncanonical ubiquitination.
Nat Commun, 11, 2020
|
|
2VWE
 
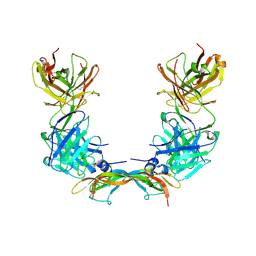 | | Crystal Structure of Vascular Endothelial Growth Factor-B in Complex with a Neutralizing Antibody Fab Fragment | | Descriptor: | ANTI-VEGF-B MONOCLONAL ANTIBODY, VASCULAR ENDOTHELIAL GROWTH FACTOR B | | Authors: | Leonard, P, Scotney, P.D, Jabeen, T, Iyer, S, Fabri, L.J, Nash, A.D, Acharya, K.R. | | Deposit date: | 2008-06-23 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of Vascular Endothelial Growth Factor-B in Complex with a Neutralising Antibody Fab Fragment.
J.Mol.Biol., 384, 2008
|
|
2WK5
 
 | | Structural features of native human thymidine phosphorylase and in complex with 5-iodouracil | | Descriptor: | GLYCEROL, THYMIDINE PHOSPHORYLASE | | Authors: | Mitsiki, E, Papageorgiou, A.C, Iyer, S, Thiyagarajan, N, Prior, S.H, Sleep, D, Finnis, C, Acharya, K.R. | | Deposit date: | 2009-06-05 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structures of Native Human Thymidine Phosphorylase and in Complex with 5-Iodouracil.
Biochem.Biophys.Res.Commun., 386, 2009
|
|
2WK6
 
 | | Structural features of native human thymidine phosphorylase and in complex with 5-iodouracil | | Descriptor: | 5-IODOURACIL, THYMIDINE PHOSPHORYLASE | | Authors: | Mitsiki, E, Papageorgiou, A.C, Iyer, S, Thiyagarajan, N, Prior, S.H, Sleep, D, Finnis, C, Acharya, K.R. | | Deposit date: | 2009-06-05 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Native Human Thymidine Phosphorylase and in Complex with 5-Iodouracil.
Biochem.Biophys.Res.Commun., 386, 2009
|
|
6UMP
 
 | | Crystal structure of MavC in complex with substrate mimic in P65 space group | | Descriptor: | MavC, Ubiquitin, Ubiquitin-conjugating enzyme E2 N | | Authors: | Puvar, K, Iyer, S, Luo, Z.Q, Das, C. | | Deposit date: | 2019-10-10 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Legionella effector MavC targets the Ube2N~Ub conjugate for noncanonical ubiquitination.
Nat Commun, 11, 2020
|
|
6E9D
 
 | | Sub-2 Angstrom Ewald Curvature-Corrected Single-Particle Cryo-EM Reconstruction of AAV-2 L336C | | Descriptor: | Capsid protein VP1 | | Authors: | Tan, Y.Z, Aiyer, S, Mietzsch, M, Hull, J.A, McKenna, R, Baker, T.S, Agbandje-McKenna, M, Lyumkis, D. | | Deposit date: | 2018-07-31 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (1.86 Å) | | Cite: | Sub-2 angstrom Ewald curvature corrected structure of an AAV2 capsid variant.
Nat Commun, 9, 2018
|
|
