2ZZM
 
 | | The complex structure of aTrm5 and tRNALeu | | Descriptor: | MAGNESIUM ION, RNA (84-MER), S-ADENOSYLMETHIONINE, ... | | Authors: | Goto-Ito, S, Ito, T, Yokoyama, S. | | Deposit date: | 2009-02-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation
Nat.Struct.Mol.Biol., 16, 2009
|
|
3A98
 
 | | Crystal structure of the complex of the interacting regions of DOCK2 and ELMO1 | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1 | | Authors: | Hanawa-Suetsugu, K, Kukimoto-Niino, M, Sekine, S, Ito, T, Mishima-Tsumagari, C, Terada, T, Shirouzu, M, Fukui, Y, Yokoyama, S. | | Deposit date: | 2009-10-21 | | Release date: | 2010-10-27 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3AY0
 
 | | Crystal structure of Methanocaldococcus jannaschii Trm5 in complex with adenosine | | Descriptor: | ADENOSINE, Uncharacterized protein MJ0883, ZINC ION | | Authors: | Goto-Ito, S, Ito, T, Hou, Y.M, Yokoyama, S. | | Deposit date: | 2011-04-21 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Differentiating analogous tRNA methyltransferases by fragments of the methyl donor.
Rna, 17, 2011
|
|
3AXZ
 
 | | Crystal structure of Haemophilus influenzae TrmD in complex with adenosine | | Descriptor: | ADENOSINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Yoshida, K, Goto-Ito, S, Ito, T, Hou, Y.M, Yokoyama, S. | | Deposit date: | 2011-04-21 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Differentiating analogous tRNA methyltransferases by fragments of the methyl donor.
Rna, 17, 2011
|
|
5YE4
 
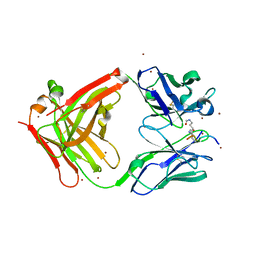 | | Crystal structure of the complex of di-acetylated histone H4 and 1A9D7 Fab fragment | | Descriptor: | 1A9D7 L chain, 1A9D7 VH CH1 chain, ZINC ION, ... | | Authors: | Matsuda, T, Ito, T, Wakamori, M, Umehara, T. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics, 13, 2018
|
|
5YE3
 
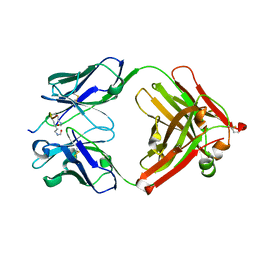 | | Crystal structure of the complex of di-acetylated histone H4 and 2A7D9 Fab fragment | | Descriptor: | 2A7D9 L chain, 2A7D9 VH CH1 chain, di-acetylated histone H4 | | Authors: | Matsuda, T, Ito, T, Wakamori, M, Umehara, T. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics, 13, 2018
|
|
6IP6
 
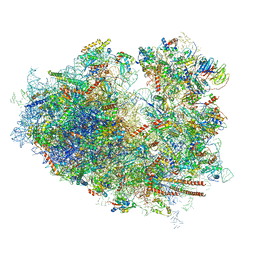 | | Cryo-EM structure of the CMV-stalled human 80S ribosome with HCV IRES (Structure iii) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6IP8
 
 | | Cryo-EM structure of the HCV IRES dependently initiated CMV-stalled 80S ribosome (Structure iv) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6IP5
 
 | | Cryo-EM structure of the CMV-stalled human 80S ribosome (Structure ii) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
5XWD
 
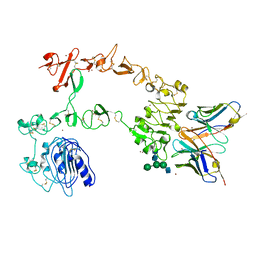 | | Crystal structure of the complex of 059-152-Fv and EGFR-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Matsuda, T, Ito, T, Shirouzu, M. | | Deposit date: | 2017-06-29 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Cell-free synthesis of functional antibody fragments to provide a structural basis for antibody-antigen interaction
PLoS ONE, 13, 2018
|
|
5ZC9
 
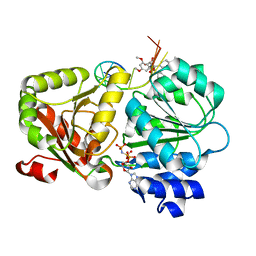 | | Crystal structure of the human eIF4A1-ATP analog-RocA-polypurine RNA complex | | Descriptor: | (1R,2R,3S,3aR,8bS)-6,8-dimethoxy-3a-(4-methoxyphenyl)-N,N-dimethyl-1,8b-bis(oxidanyl)-3-phenyl-2,3-dihydro-1H-cyclopenta[b][1]benzofuran-2-carboxamide, Eukaryotic initiation factor 4A-I, MAGNESIUM ION, ... | | Authors: | Iwasaki, W, Takahashi, M, Sakamoto, A, Iwasaki, S, Ito, T. | | Deposit date: | 2018-02-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Translation Inhibitor Rocaglamide Targets a Bimolecular Cavity between eIF4A and Polypurine RNA.
Mol. Cell, 73, 2019
|
|
6K72
 
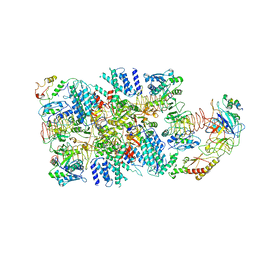 | | eIF2(aP) - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Kashiwagi, K, Yokoyama, T, Ito, T. | | Deposit date: | 2019-06-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
6K71
 
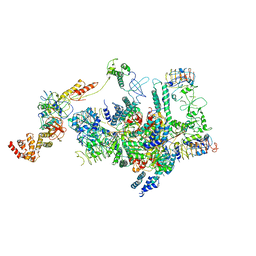 | | eIF2 - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Kashiwagi, K, Yokoyama, T, Ito, T. | | Deposit date: | 2019-06-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
7CPJ
 
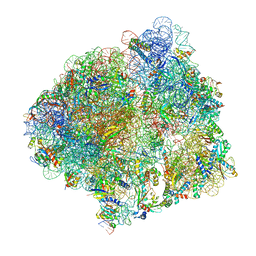 | | ycbZ-stalled 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shirouzu, M, Ito, T. | | Deposit date: | 2020-08-07 | | Release date: | 2021-12-08 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The landscape of translational stall sites in bacteria revealed by monosome and disome profiling.
Rna, 28, 2022
|
|
2DPC
 
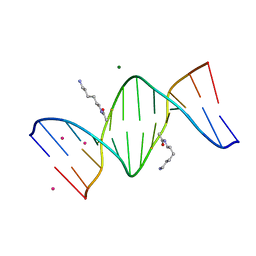 | | Crystal Structure of d(CGCGAATXCGCG) Where X is 5-(N-aminohexyl)carbamoyl-2'-O-methyluridine | | Descriptor: | (6-AMINOHEXYL)CARBAMIC ACID, COBALT (II) ION, DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*(OMU)P*DCP*DGP*DCP*DG)-3'), ... | | Authors: | Juan, E.C.M, Kondo, J, Kurihara, T, Ito, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2006-05-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of DNA:DNA and DNA:RNA duplexes containing 5-(N-aminohexyl)carbamoyl-modified uracils reveal the basis for properties as antigene and antisense molecules
Nucleic Acids Res., 35, 2007
|
|
2DP7
 
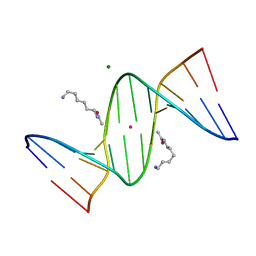 | | Crystal Structure of D(CGCGAATXCGCG) Where X is 5-(N-aminohexyl)carbamoyl-2'-deoxyuridine | | Descriptor: | (6-AMINOHEXYL)CARBAMIC ACID, DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*DUP*DCP*DGP*DCP*DG)-3'), MAGNESIUM ION, ... | | Authors: | Juan, E.C.M, Kondo, J, Kurihara, T, Ito, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2006-05-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of DNA:DNA and DNA:RNA duplexes containing 5-(N-aminohexyl)carbamoyl-modified uracils reveal the basis for properties as antigene and antisense molecules
Nucleic Acids Res., 35, 2007
|
|
2DPB
 
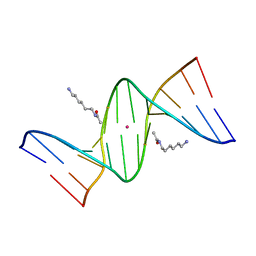 | | Crystal Structure of d(CGCGAATXCGCG) Where X is 5-(N-aminohexyl)carbamoyl-2'-deoxyuridine | | Descriptor: | (6-AMINOHEXYL)CARBAMIC ACID, DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*DUP*DCP*DGP*DCP*DG)-3'), POTASSIUM ION | | Authors: | Juan, E.C.M, Kondo, J, Kurihara, T, Ito, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2006-05-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of DNA:DNA and DNA:RNA duplexes containing 5-(N-aminohexyl)carbamoyl-modified uracils reveal the basis for properties as antigene and antisense molecules
Nucleic Acids Res., 35, 2007
|
|
6JLZ
 
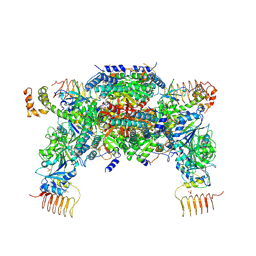 | | P-eIF2a - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, PHOSPHATE ION, Probable translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-01 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
6JLY
 
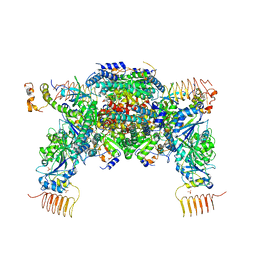 | | eIF2a - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, PHOSPHATE ION, Probable translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
2DQQ
 
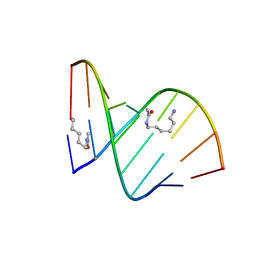 | | Crystal Structure of d(CXCTXCTTC):r(gaagaagag) Where X is 5-(N-aminohexyl)carbamoyl-2'-O-methyluridine | | Descriptor: | (6-AMINOHEXYL)CARBAMIC ACID, DNA (5'-D(*DCP*(OMU)P*DCP*DTP*(OMU)P*DCP*DTP*DTP*DC)-3'), RNA (5'-R(*GP*AP*AP*GP*AP*AP*GP*AP*G)-3') | | Authors: | Juan, E.C.M, Kondo, J, Ito, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2006-05-29 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of DNA:DNA and DNA:RNA duplexes containing 5-(N-aminohexyl)carbamoyl-modified uracils reveal the basis for properties as antigene and antisense molecules
Nucleic Acids Res., 35, 2007
|
|
2DQP
 
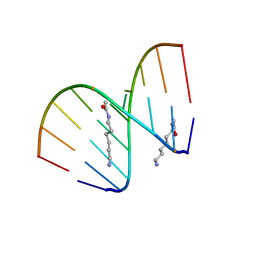 | | Structural analyses of DNA:DNA and RNA:DNA duplexes containing 5-(N-aminohexyl)carbamoyl modified uridines | | Descriptor: | (6-AMINOHEXYL)CARBAMIC ACID, DNA (5'-D(*DCP*(OMU)P*DCP*DTP*(OMU)P*DCP*DTP*DTP*DC)-3'), RNA (5'-R(*GP*AP*AP*GP*AP*AP*GP*AP*G)-3') | | Authors: | Juan, E.C.M, Kondo, J, Ito, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2006-05-29 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of DNA:DNA and DNA:RNA duplexes containing 5-(N-aminohexyl)carbamoyl-modified uracils reveal the basis for properties as antigene and antisense molecules
Nucleic Acids Res., 35, 2007
|
|
2DQO
 
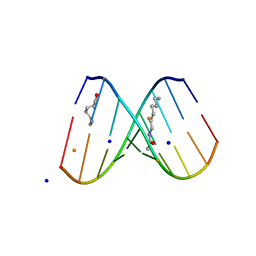 | | Crystal Structure of d(CXCTXCTTC):r(gaagaagag) Where X is 5-(N-aminohexyl)carbamoyl-2'-O-methyluridine | | Descriptor: | (6-AMINOHEXYL)CARBAMIC ACID, BARIUM ION, DNA (5'-D(*DCP*(OMU)P*DCP*DTP*(OMU)P*DCP*DTP*DTP*DC)-3'), ... | | Authors: | Juan, E.C.M, Kondo, J, Ito, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2006-05-29 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of DNA:DNA and DNA:RNA duplexes containing 5-(N-aminohexyl)carbamoyl-modified uracils reveal the basis for properties as antigene and antisense molecules
Nucleic Acids Res., 35, 2007
|
|
1TZ1
 
 | | Solution structure of the PB1 domain of CDC24P (short form) | | Descriptor: | Cell division control protein 24 | | Authors: | Yoshinaga, S, Terasawa, H, Ogura, K, Noda, Y, Ito, T, Sumimoto, H, Inagaki, F. | | Deposit date: | 2004-07-09 | | Release date: | 2005-09-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PB1 domain of CDC24P (short form)
To be Published
|
|
3VSS
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain complexed with fructose | | Descriptor: | Beta-fructofuranosidase, beta-D-fructofuranose | | Authors: | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
3VSR
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain | | Descriptor: | Beta-fructofuranosidase | | Authors: | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
