6XBZ
 
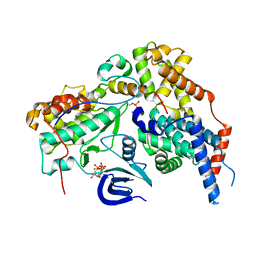 | | Structure of the human CDK-activating kinase | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Greber, B.J, Perez-Bertoldi, J.M, Lim, K, Iavarone, A.T, Toso, D.B, Nogales, E. | | Deposit date: | 2020-06-07 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The cryoelectron microscopy structure of the human CDK-activating kinase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XD3
 
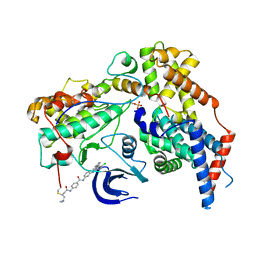 | | Structure of the human CAK in complex with THZ1 | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Greber, B.J, Perez-Bertoldi, J.M, Lim, K, Iavarone, A.T, Toso, D.B, Nogales, E. | | Deposit date: | 2020-06-09 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The cryoelectron microscopy structure of the human CDK-activating kinase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4CMQ
 
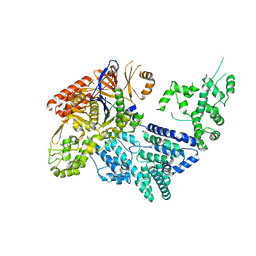 | | Crystal structure of Mn-bound S.pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MANGANESE (II) ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-17 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA- Mediated Conformational Activation
Science, 343, 2014
|
|
4CMP
 
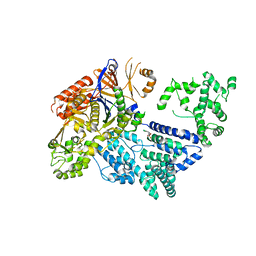 | | Crystal structure of S. pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation.
Science, 343, 2014
|
|
7LAF
 
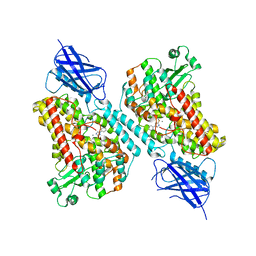 | | 15-lipoxygenase-2 loop mutant bound to imidazole-based inhibitor | | Descriptor: | 3-{[(4-methylphenyl)methyl]sulfanyl}-1-phenyl-1H-1,2,4-triazole, MANGANESE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX15B | | Authors: | Newcomer, M.E, Gilbert, N.C, Neau, D.B. | | Deposit date: | 2021-01-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Kinetic and structural investigations of novel inhibitors of human epithelial 15-lipoxygenase-2.
Bioorg.Med.Chem., 46, 2021
|
|
8VIY
 
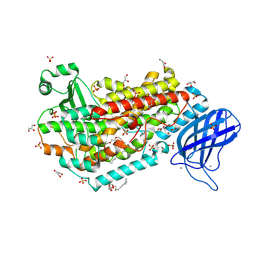 | | 15-Lipoxygenase-2 V427L | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, FE (II) ION, ... | | Authors: | Gilbert, N.C, Offenbacher, A.R, Ohler, A.R.L. | | Deposit date: | 2024-01-05 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Identification of the Thermal Activation Network in Human 15-Lipoxygenase-2: Divergence from Plant Orthologs and Its Relationship to Hydrogen Tunneling Activation Barriers.
Acs Catalysis, 14, 2024
|
|
3SBO
 
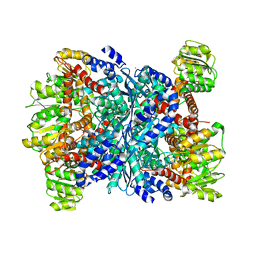 | | Structure of E.coli GDH from native source | | Descriptor: | CHLORIDE ION, NADP-specific glutamate dehydrogenase | | Authors: | Gee, C.L, Zubieta, C, Echols, N, Totir, M. | | Deposit date: | 2011-06-06 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
2XHY
 
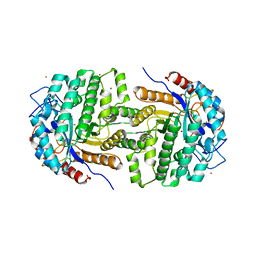 | | Crystal Structure of E.coli BglA | | Descriptor: | 6-PHOSPHO-BETA-GLUCOSIDASE BGLA, BROMIDE ION, SULFATE ION | | Authors: | Totir, M, Zubieta, C, Echols, N, May, A.P, Gee, C.L, nanao, M, alber, T. | | Deposit date: | 2010-06-24 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
9MHG
 
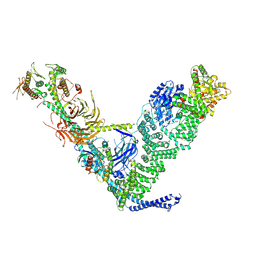 | |
9MHH
 
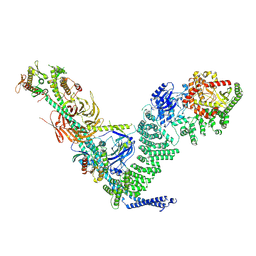 | |
9MHF
 
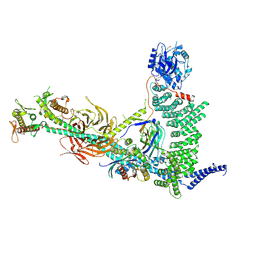 | |
5T5V
 
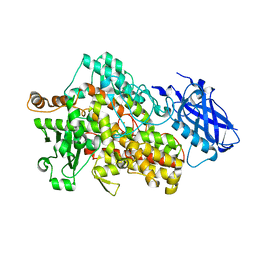 | | LIPOXYGENASE-1 (SOYBEAN) AT 293K | | Descriptor: | FE (III) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrogen-Deuterium Exchange of Lipoxygenase Uncovers a Relationship between Distal, Solvent Exposed Protein Motions and the Thermal Activation Barrier for Catalytic Proton-Coupled Electron Tunneling.
ACS Cent Sci, 3, 2017
|
|
6E13
 
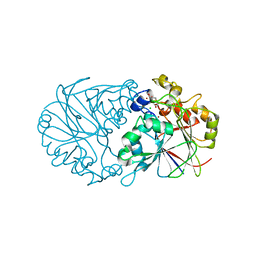 | | Pseudomonas putida PqqB with a non-physiological zinc at the active site binds the substrate mimic, 5-cysteinyl-3,4-dihydroxyphenylalanine (5-Cys-DOPA), non-specifically but supports the proposed function of the enzyme in pyrroloquinoline quinone biosynthesis. | | Descriptor: | 3-{[(2S)-2-amino-2-carboxyethyl]sulfanyl}-5-hydroxy-L-tyrosine, CHLORIDE ION, Coenzyme PQQ synthesis protein B, ... | | Authors: | Evans III, R.L, Wilmot, C.M. | | Deposit date: | 2018-07-09 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Discovery of Hydroxylase Activity for PqqB Provides a Missing Link in the Pyrroloquinoline Quinone Biosynthetic Pathway.
J.Am.Chem.Soc., 141, 2019
|
|
5TQP
 
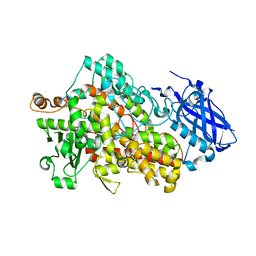 | | LIPOXYGENASE-1 (SOYBEAN) I553G MUTANT AT 300K | | Descriptor: | FE (III) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen-Deuterium Exchange of Lipoxygenase Uncovers a Relationship between Distal, Solvent Exposed Protein Motions and the Thermal Activation Barrier for Catalytic Proton-Coupled Electron Tunneling.
ACS Cent Sci, 3, 2017
|
|
3NBU
 
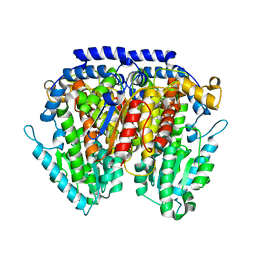 | | Crystal structure of pGI glucosephosphate isomerase | | Descriptor: | CHLORIDE ION, Glucose-6-phosphate isomerase | | Authors: | Alber, T, Zubieta, C, Totir, M, May, A, Echols, N. | | Deposit date: | 2010-06-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
3N6Q
 
 | | Crystal structure of YghZ from E. coli | | Descriptor: | MAGNESIUM ION, YghZ aldo-keto reductase | | Authors: | Zubieta, C, Totir, M, Echols, N, May, A, Alber, T. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
6MEM
 
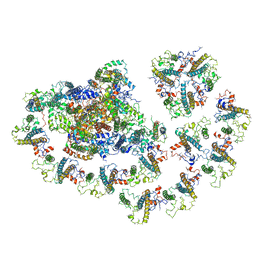 | |
3F61
 
 | |
3F69
 
 | | Crystal structure of the Mycobacterium tuberculosis PknB mutant kinase domain in complex with KT5720 | | Descriptor: | SULFATE ION, Serine/threonine-protein kinase pknB, hexyl (5S,6R,8R)-6-hydroxy-5-methyl-13-oxo-5,6,7,8-tetrahydro-13H-5,8-epoxy-4b,8a,14-triazadibenzo[b,h]cycloocta[1,2,3,4-jkl]c yclopenta[e]-as-indacene-6-carboxylate | | Authors: | Alber, T, Mieczkowski, C.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Auto-activation mechanism of the Mycobacterium tuberculosis PknB receptor Ser/Thr kinase.
Embo J., 27, 2008
|
|
7SOJ
 
 | |
7SOI
 
 | | Structure of I552A Soybean Lipoxygenase at 277K | | Descriptor: | FE (III) ION, Lipoxygenase, SODIUM ION | | Authors: | Gee, C.L, Offenbacher, A.R, Hu, S. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Temporal and spatial resolution of distal protein motions that activate hydrogen tunneling in soybean lipoxygenase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4OGC
 
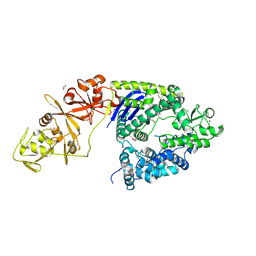 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | Descriptor: | ACETATE ION, HNH endonuclease domain protein, MAGNESIUM ION, ... | | Authors: | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
4QSK
 
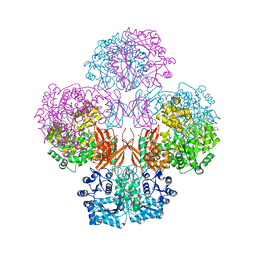 | | Crystal Structure of L. monocytogenes Pyruvate Carboxylase in complex with Cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CITRATE ANION, MANGANESE (II) ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2014-07-04 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Cyclic Dinucleotide c-di-AMP Is an Allosteric Regulator of Metabolic Enzyme Function.
Cell(Cambridge,Mass.), 158, 2014
|
|
4QSH
 
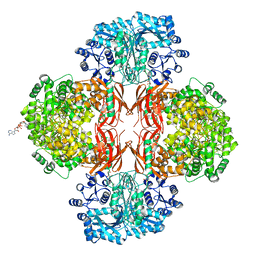 | | Crystal Structure of L. monocytogenes Pyruvate Carboxylase in complex with Cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CITRATE ANION, MANGANESE (II) ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2014-07-04 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The Cyclic Dinucleotide c-di-AMP Is an Allosteric Regulator of Metabolic Enzyme Function.
Cell(Cambridge,Mass.), 158, 2014
|
|
4OGE
 
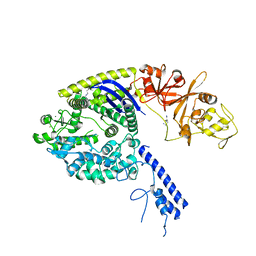 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | Descriptor: | HNH endonuclease domain protein, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
