2OK3
 
 | | X-ray structure of human cyclophilin J at 2.0 angstrom | | Descriptor: | NICKEL (II) ION, Peptidyl-prolyl cis-trans isomerase-like 3 | | Authors: | Xia, Z. | | Deposit date: | 2007-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Cyclophilin J, a novel peptidyl-prolyl isomerase, can induce cellular G1/S arrest and repress the growth of Hepatocellular carcinoma
To be Published
|
|
4HGT
 
 | | Crystal structure of ck1d with compound 13 | | Descriptor: | 2-{2-[(3,4-difluorophenoxy)methyl]-5-methoxypyridin-4-yl}-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Casein kinase I isoform delta | | Authors: | Huang, X. | | Deposit date: | 2012-10-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of Potent and Selective CK1 gamma Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
4HGL
 
 | | Crystal structure of ck1g3 with compound 1 | | Descriptor: | 2-[5-methoxy-2-(quinolin-3-yl)pyrimidin-4-yl]-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Casein kinase I isoform gamma-3, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2012-10-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Potent and Selective CK1 gamma Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
4MD6
 
 | | Crystal structure of PDE5 in complex with inhibitor 5R | | Descriptor: | 3-(4-hydroxybenzyl)-1-(thiophen-2-yl)chromeno[2,3-c]pyrrol-9(2H)-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Cui, W, Huang, M, Shao, Y, Luo, H. | | Deposit date: | 2013-08-22 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 3-(4-hydroxybenzyl)-1-(thiophen-2-yl)chromeno[2,3-c]pyrrol-9(2H)-one as a phosphodiesterase-5 inhibitor and its complex crystal structure.
Biochem Pharmacol, 89, 2014
|
|
5WUK
 
 | |
4MSK
 
 | | Co-crystal structure of tankyrase 1 with compound 34 | | Descriptor: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[4-(5-phenyl-1,3,4-oxadiazol-2-yl)phenyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4MT9
 
 | | Co-crystal structure of tankyrase 1 with compound 49 | | Descriptor: | N-[trans-4-(4-cyanophenoxy)cyclohexyl]-3-[(4-oxo-3,4-dihydroquinazolin-2-yl)sulfanyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-19 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4MSG
 
 | | Crystal structure of tankyrase 1 with compound 22 | | Descriptor: | 3-[(4-oxo-3,4-dihydroquinazolin-2-yl)sulfanyl]-N-[trans-4-(5-phenyl-1,3,4-oxadiazol-2-yl)cyclohexyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
5CHL
 
 | | Structural basis of H2A.Z recognition by YL1 histone chaperone component of SRCAP/SWR1 chromatin remodeling complex | | Descriptor: | Histone H2A.Z, Vacuolar protein sorting-associated protein 72 homolog | | Authors: | Shan, S, Liang, X, Pan, L, Wu, C, Zhou, Z. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1
Nat.Struct.Mol.Biol., 23, 2016
|
|
7YQF
 
 | | Crystal structure of PDE4D complexed with glycyrrhisoflavone | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-4,5-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YSX
 
 | | Crystal structure of PDE4D complexed with licoisoflavone A | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-2,4-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1F4V
 
 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY BOUND TO THE N-TERMINUS OF FLIM | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, FLAGELLAR MOTOR SWITCH PROTEIN, ... | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Henderson, R.K, King, D, Huang, L.S, Kustu, S, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-06-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of an activated response regulator bound to its target.
Nat.Struct.Biol., 8, 2001
|
|
2XTJ
 
 | | The crystal structure of PCSK9 in complex with 1D05 Fab | | Descriptor: | CALCIUM ION, FAB FROM A HUMAN MONOCLONAL ANTIBODY, 1D05, ... | | Authors: | Di Marco, S, Volpari, C, Carfi, A. | | Deposit date: | 2010-10-10 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Pcsk9-Binding Antibody that Structurally Mimics the Egf(A) Domain of Ldl-Receptor Reduces Ldl Cholesterol in Vivo.
J.Lipid Res., 52, 2011
|
|
8U7U
 
 | | Proteasome 20S Core Particle from Beta 3 D205 deletion | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Velez, B, Blickling, M, Razi, A, Hanna, J. | | Deposit date: | 2023-09-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Mechanism of autocatalytic activation during proteasome assembly.
Nat.Struct.Mol.Biol., 2024
|
|
8U6Y
 
 | | Preholo-Proteasome from Beta 3 D205 deletion | | Descriptor: | Proteasome assembly chaperone 2, Proteasome chaperone 1, Proteasome maturation factor UMP1, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Velez, B, Blickling, M, Razi, A, Hanna, J. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of autocatalytic activation during proteasome assembly.
Nat.Struct.Mol.Biol., 2024
|
|
4RWB
 
 | | Racemic influenza M2-TM crystallized from monoolein lipidic cubic phase | | Descriptor: | Matrix protein 2, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Mortenson, D.E, Steinkruger, J.D, Kreitler, D.F, Gellman, S.H, Forest, K.T. | | Deposit date: | 2014-12-02 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of a heterochiral coiled coil.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RWC
 
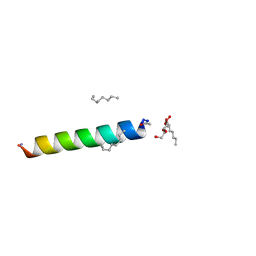 | | Racemic M2-TM crystallized from racemic detergent | | Descriptor: | Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Mortenson, D.E, Steinkruger, J.D, Kreitler, D.F, Gellman, S.H, Forest, K.T. | | Deposit date: | 2014-12-02 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | High-resolution structures of a heterochiral coiled coil.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1Y9X
 
 | |
5DH3
 
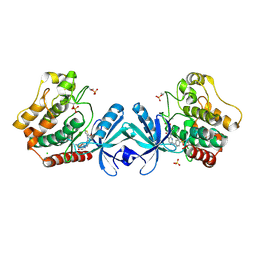 | | Crystal structure of MST2 in complex with XMU-MP-1 | | Descriptor: | 4-[(5,10-dimethyl-6-oxo-6,10-dihydro-5H-pyrimido[5,4-b]thieno[3,2-e][1,4]diazepin-2-yl)amino]benzenesulfonamide, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2015-08-29 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration
Sci Transl Med, 8, 2016
|
|
6JQH
 
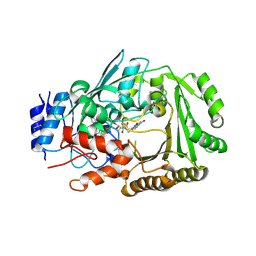 | | Crystal structure of MaDA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MaDA | | Authors: | Du, X.X, Lei, X.G. | | Deposit date: | 2019-03-31 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | FAD-dependent enzyme-catalysed intermolecular [4+2] cycloaddition in natural product biosynthesis.
Nat.Chem., 12, 2020
|
|
5BPP
 
 | | Structure of human Leukotriene A4 hydrolase in complex with inhibitor 4AZ | | Descriptor: | 2-(4-butoxyphenyl)-N-hydroxyacetamide, ACETATE ION, Leukotriene A-4 hydrolase, ... | | Authors: | Huang, J, Dong, N.N, Xiao, Q, Ou, P.Y, Wu, D, Lu, W.Q. | | Deposit date: | 2015-05-28 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Bufexamac ameliorates LPS-induced acute lung injury in mice by targeting LTA4H
Sci Rep, 6, 2016
|
|
7ML7
 
 | |
1S13
 
 | | Human Heme Oxygenase Oxidatition of alpha- and gamma-meso-Phenylhemes | | Descriptor: | 2-PHENYLHEME, Heme oxygenase 1 | | Authors: | Wang, J, Niemevz, F, Lad, L, Buldain, G, Poulos, T.L, Ortiz de Montellano, P.R. | | Deposit date: | 2004-01-05 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Human heme oxygenase oxidation of 5- and 15-phenylhemes.
J.Biol.Chem., 279, 2004
|
|
1T5P
 
 | | Human Heme Oxygenase Oxidation of alpha- and gamma-meso-phenylhemes | | Descriptor: | 12-PHENYLHEME, Heme oxygenase 1 | | Authors: | Wang, J, Niemevz, F, Lad, L, Buldain, G, Poulos, T.L, Ortiz de Montellano, P.R. | | Deposit date: | 2004-05-05 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Human heme oxygenase oxidation of 5- and 15-phenylhemes.
J.Biol.Chem., 279, 2004
|
|
5YM3
 
 | | CYP76AH1-4pi from salvia miltiorrhiza | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Ferruginol synthase, MANGANESE (II) ION, ... | | Authors: | Chang, Z. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of CYP76AH1 in 4-PI-bound state from Salvia miltiorrhiza.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
