2R7U
 
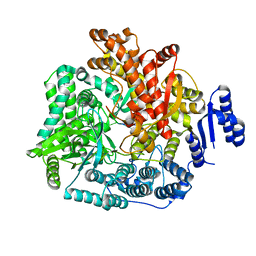 | | Crystal Structure of Rotavirus SA11 VP1/RNA (AAAAGCC) Complex | | Descriptor: | RNA (5'-R(*AP*A*AP*AP*GP*CP*C)-3'), RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
2R7X
 
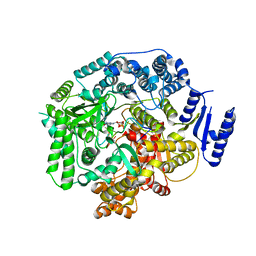 | | Crystal Structure of Rotavirus SA11 VP1/RNA (UGUGACC)/GTP complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA (5'-R(*UP*GP*UP*GP*AP*CP*C)-3'), RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
2R7W
 
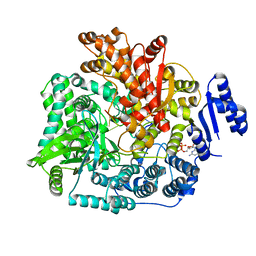 | | Crystal Structure of Rotavirus SA11 VP1/RNA (UGUGACC)/mRNA 5'-CAP (m7GpppG) complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA (5'-R(*UP*GP*UP*GP*AP*CP*C)-3'), RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
2SPC
 
 | | CRYSTAL STRUCTURE OF THE REPETITIVE SEGMENTS OF SPECTRIN | | Descriptor: | SPECTRIN | | Authors: | Yan, Y, Winograd, E, Viel, A, Cronin, T, Harrison, S.C, Branton, D. | | Deposit date: | 1994-03-01 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the repetitive segments of spectrin.
Science, 262, 1993
|
|
2SRC
 
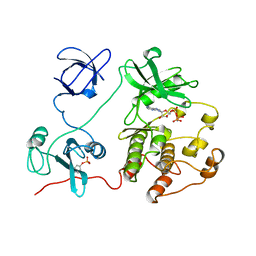 | | CRYSTAL STRUCTURE OF HUMAN TYROSINE-PROTEIN KINASE C-SRC, IN COMPLEX WITH AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, TYROSINE-PROTEIN KINASE SRC | | Authors: | Xu, W, Doshi, A, Lei, M, Eck, M.J, Harrison, S.C. | | Deposit date: | 1998-12-29 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of c-Src reveal features of its autoinhibitory mechanism.
Mol.Cell, 3, 1999
|
|
4F5X
 
 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | Descriptor: | Intermediate capsid protein VP6, RNA-directed RNA polymerase, VP2 protein, ... | | Authors: | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2012-05-13 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Location of the dsRNA-Dependent Polymerase, VP1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
3ML6
 
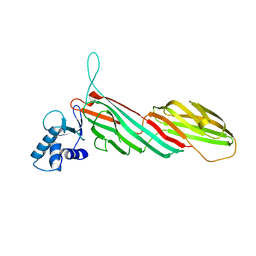 | | a complex between Dishevelled2 and clathrin adaptor AP-2 | | Descriptor: | Chimeric complex between protein Dishevelled2 homolog dvl-2 and clathrin adaptor AP-2 complex subunit mu | | Authors: | Yu, A, Xing, Y, Harrison, S.C, Kirchhausen, T.L. | | Deposit date: | 2010-04-16 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural analysis of the interaction between Dishevelled2 and clathrin AP-2 adaptor, a critical step in noncanonical Wnt signaling.
Structure, 18, 2010
|
|
3N4S
 
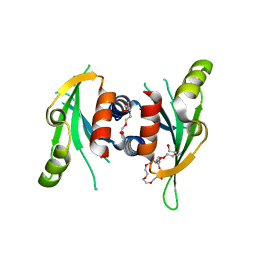 | | Structure of Csm1 C-terminal domain, P21212 form | | Descriptor: | Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N7N
 
 | | Structure of Csm1/Lrs4 complex | | Descriptor: | Monopolin complex subunit CSM1, Monopolin complex subunit LRS4 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N4R
 
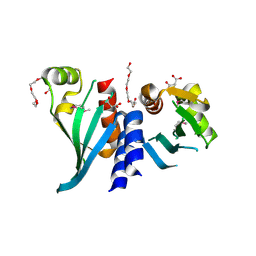 | | Structure of Csm1 C-terminal domain, R3 form | | Descriptor: | MALONATE ION, Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N4X
 
 | | Structure of Csm1 full-length | | Descriptor: | Monopolin complex subunit CSM1 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-23 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
4GSX
 
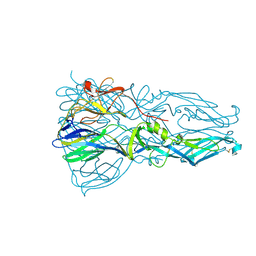 | | High resolution structure of dengue virus serotype 1 sE containing stem | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Klein, D.E, Choi, J.L, Harrison, S.C. | | Deposit date: | 2012-08-28 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structure of a dengue virus envelope protein late-stage fusion intermediate.
J.Virol., 87, 2013
|
|
4GT0
 
 | | Structure of dengue virus serotype 1 sE containing stem to residue 421 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Klein, D.E, Choi, J.L, Harrison, S.C. | | Deposit date: | 2012-08-28 | | Release date: | 2012-12-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of a dengue virus envelope protein late-stage fusion intermediate.
J.Virol., 87, 2013
|
|
4HKB
 
 | | CH67 Fab (unbound) from the CH65-67 Lineage | | Descriptor: | CH67 heavy chain, CH67 light chain | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-15 | | Release date: | 2012-11-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HK3
 
 | | I2 Fab (unbound) from CH65-CH67 Lineage | | Descriptor: | I2 heavy chain, I2 light chain | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-14 | | Release date: | 2012-11-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HKX
 
 | | Influenza hemagglutinin in complex with CH67 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH67 heavy chain, CH67 light chain, ... | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-15 | | Release date: | 2012-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HK0
 
 | | UCA Fab (unbound) from CH65-CH67 Lineage | | Descriptor: | UCA heavy chain, UCA light chain | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-14 | | Release date: | 2012-11-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IT3
 
 | | Crystal Structure of Iml3 from S. cerevisiae | | Descriptor: | Central kinetochore subunit IML3 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2013-01-17 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | An iml3-chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation.
Cell Rep, 5, 2013
|
|
4JE3
 
 | |
3ZX8
 
 | | Cryo-EM reconstruction of native and expanded Turnip Crinkle virus | | Descriptor: | CAPSID PROTEIN | | Authors: | Bakker, S.E, Robottom, J, Hogle, J.M, Maeda, A, Pearson, A.R, Stockley, P.G, Ranson, N.A, Harrison, S.C. | | Deposit date: | 2011-08-08 | | Release date: | 2012-07-18 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Isolation of an Asymmetric RNA Uncoating Intermediate for a Single-Stranded RNA Plant Virus.
J.Mol.Biol., 417, 2012
|
|
7UHZ
 
 | | Post-fusion ectodomain of HSV-1 gB in complex with BMPC-23 Fab | | Descriptor: | BMPC-23 Fab Heavy chain, BMPC-23 Fab Light chain, Envelope glycoprotein B | | Authors: | Windsor, I.W, Kong, S.L, Garforth, S.J, Almo, S.C, Harrison, S.C. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice.
J.Clin.Invest., 133, 2023
|
|
7UI0
 
 | | Post-fusion ectodomain of HSV-1 gB in complex with HSV010-13 Fab | | Descriptor: | Envelope glycoprotein B, HSV10-13 Fab Heavy chain, HSV10-13 Light chain | | Authors: | Windsor, I.W, Kong, S.L, Garforth, S.J, Almo, S.C, Harrison, S.C. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice.
J.Clin.Invest., 133, 2023
|
|
5LSJ
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C delta-HEAD2 COMPLEX | | Descriptor: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
5LSI
 
 | | CRYSTAL STRUCTURE OF THE KINETOCHORE MIS12 COMPLEX HEAD2 SUBDOMAIN CONTAINING DSN1 AND NSL1 FRAGMENTS | | Descriptor: | Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, SULFATE ION | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
5LSK
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C COMPLEX | | Descriptor: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
