3F7T
 
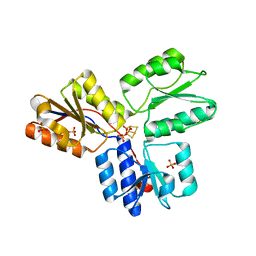 | | Structure of active IspH shows a novel fold with a [3Fe-4S] cluster in the catalytic centre | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, PHOSPHATE ION, ... | | Authors: | Graewert, T, Eppinger, J, Rohdich, F, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2008-11-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of active IspH enzyme from Escherichia coli provides mechanistic insights into substrate reduction.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3UNE
 
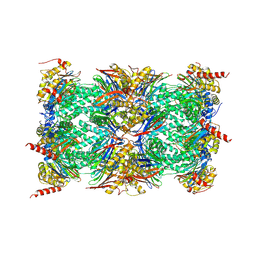 | | Mouse constitutive 20S proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
3UNH
 
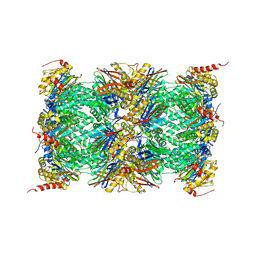 | | Mouse 20S immunoproteasome | | Descriptor: | CHLORIDE ION, IODIDE ION, POTASSIUM ION, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
3UN4
 
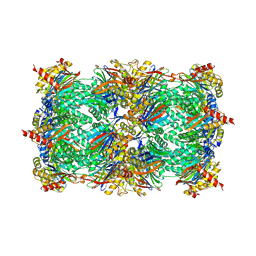 | | Yeast 20S proteasome in complex with PR-957 (morpholine) | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, Proteasome component C1, Proteasome component C11, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
3UNF
 
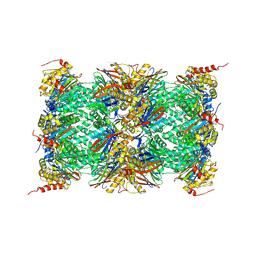 | | Mouse 20S immunoproteasome in complex with PR-957 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, CHLORIDE ION, IODIDE ION, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
3UNB
 
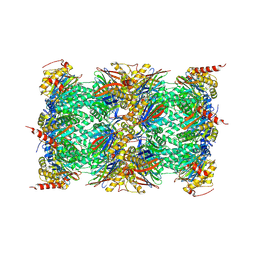 | | Mouse constitutive 20S proteasome in complex with PR-957 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
3UN8
 
 | | Yeast 20S proteasome in complex with PR-957 (epoxide) | | Descriptor: | 2-(acetylamino)-4,5-anhydro-1,2-dideoxy-4-methyl-1-phenyl-D-xylitol, Proteasome component C1, Proteasome component C11, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
6SJR
 
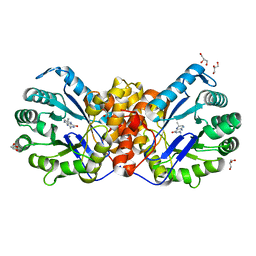 | | Methyltransferase of the MtgA N227A mutant from Desulfitobacterium hafniense in complex with tetrahydrofolate | | Descriptor: | GLYCEROL, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, Tetrahydromethanopterin S-methyltransferase | | Authors: | Badmann, T, Groll, M. | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures in Tetrahydrofolate Methylation in Desulfitobacterial Glycine Betaine Metabolism at Atomic Resolution.
Chembiochem, 21, 2020
|
|
6SK4
 
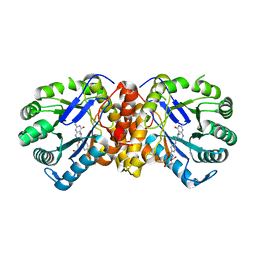 | | Methyltransferase MtgA from Desulfitobacterium hafniense in complex with methyl-tetrahydrofolate (P21) | | Descriptor: | GLYCEROL, Methylcorrinoid:tetrahydrofolate methyltransferase, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID | | Authors: | Badmann, T, Groll, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures in Tetrahydrofolate Methylation in Desulfitobacterial Glycine Betaine Metabolism at Atomic Resolution.
Chembiochem, 21, 2020
|
|
6SJ8
 
 | | Methyltransferase MtgA from Desulfitobacterium hafniense in complex with tetrahydrofolate | | Descriptor: | (6S)-2-amino-6-methyl-5,6,7,8-tetrahydropteridin-4(3H)-one, GLYCEROL, SODIUM ION, ... | | Authors: | Badmann, T, Groll, M. | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures in Tetrahydrofolate Methylation in Desulfitobacterial Glycine Betaine Metabolism at Atomic Resolution.
Chembiochem, 21, 2020
|
|
6SJK
 
 | | Methyltransferase MtgA from Desulfitobacterium hafniense | | Descriptor: | GLYCEROL, Tetrahydromethanopterin S-methyltransferase | | Authors: | Badmann, T, Groll, M. | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures in Tetrahydrofolate Methylation in Desulfitobacterial Glycine Betaine Metabolism at Atomic Resolution.
Chembiochem, 21, 2020
|
|
6SJP
 
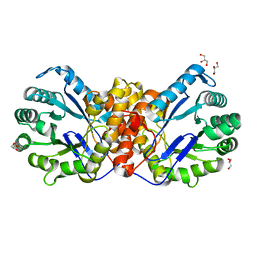 | |
6SJS
 
 | | Methyltransferase of the MtgA N227A mutant from Desulfitobacterium hafniense in complex with methyl-tetrahydrofolate | | Descriptor: | GLYCEROL, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, SODIUM ION, ... | | Authors: | Badmann, T, Groll, M. | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures in Tetrahydrofolate Methylation in Desulfitobacterial Glycine Betaine Metabolism at Atomic Resolution.
Chembiochem, 21, 2020
|
|
6TTY
 
 | | Structure of ClpP from Staphylococcus aureus (apo, closed state) | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Malik, I.T, Pereira, R, Vielberg, M.-T, Mayer, C, Straetener, J, Thomy, D, Famulla, K, Castro, H.C, Sass, P, Groll, M, Broetz-Oesterheldt, H. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional Characterisation of ClpP Mutations Conferring Resistance to Acyldepsipeptide Antibiotics in Firmicutes.
Chembiochem, 21, 2020
|
|
6TTZ
 
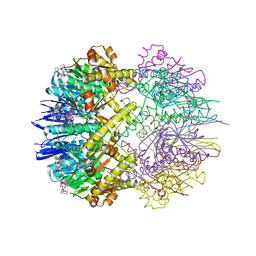 | | Structure of the ClpP:ADEP4-complex from Staphylococcus aureus (open state) | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, N-[(2S)-3-(3,5-difluorophenyl)-1-[[(3S,9S,13S,15R,19S,22S)-15,19-dimethyl-2,8,12,18,21-pentaoxo-11-oxa-1,7,17,20-tetrazatetracyclo[20.4.0.03,7.013,17]hexacosan-9-yl]amino]-1-oxopropan-2-yl]heptanamide | | Authors: | Malik, I.T, Pereira, R, Vielberg, M.-T, Mayer, C, Straetener, J, Thomy, D, Famulla, K, Castro, H.C, Sass, P, Groll, M, Broetz-Oesterheldt, H. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Characterisation of ClpP Mutations Conferring Resistance to Acyldepsipeptide Antibiotics in Firmicutes.
Chembiochem, 21, 2020
|
|
5BXL
 
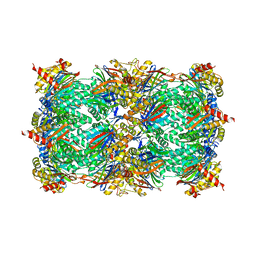 | | Yeast 20S proteasome beta2-G170A mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defective immuno- and thymoproteasome assembly causes severe immunodeficiency.
Sci Rep, 8, 2018
|
|
5BXN
 
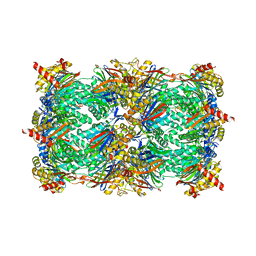 | | Yeast 20S proteasome beta2-G170A mutant in complex with Bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defective immuno- and thymoproteasome assembly causes severe immunodeficiency.
Sci Rep, 8, 2018
|
|
5NYW
 
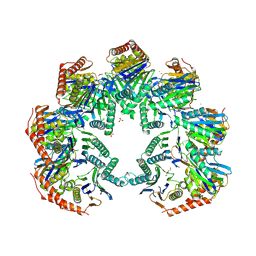 | | Anbu (ancestral beta-subunit) from Yersinia bercovieri | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | Authors: | Piasecka, A, Czapinska, H, Vielberg, M, Szczepanowski, R.H, Reed, S, Groll, M, Bochtler, M. | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | The Y. bercovieri Anbu crystal structure sheds light on the evolution of highly (pseudo)symmetric multimers.
J. Mol. Biol., 430, 2018
|
|
3SZU
 
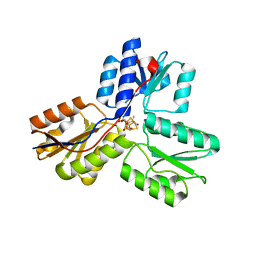 | | IspH:HMBPP complex structure of E126Q mutant | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
3T7V
 
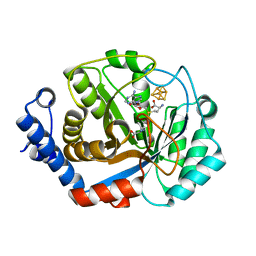 | | Crystal structure of methylornithine synthase (PylB) | | Descriptor: | 5-amino-D-isoleucine, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, ... | | Authors: | Quitterer, F, List, A, Eisenreich, W, Bacher, A, Groll, M. | | Deposit date: | 2011-07-31 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of methylornithine synthase (PylB): insights into the pyrrolysine biosynthesis.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3T0G
 
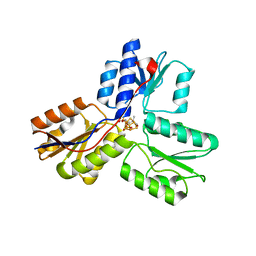 | | IspH:HMBPP (substrate) structure of the T167C mutant | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
3T0F
 
 | | IspH:HMBPP (substrate) structure of the E126D mutant | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
3SZL
 
 | | IspH:Ligand Mutants - wt 70sec | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
3SZO
 
 | | IspH:HMBPP complex after 3 minutes X-ray pre-exposure | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
3TDD
 
 | | Crystal structure of yeast CP in complex with Belactosin C | | Descriptor: | Proteasome component C1, Proteasome component C11, Proteasome component C5, ... | | Authors: | Korotkov, V.S, Ludwig, A, Larionov, O.V, Lygin, A.V, Groll, M, de Meijere, A. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthesis and biological activity of optimized belactosin C congeners.
Org.Biomol.Chem., 9, 2011
|
|
