3TND
 
 | | Crystal structure of Shigella flexneri VapBC toxin-antitoxin complex | | Descriptor: | Antitoxin VapB, SODIUM ION, SULFATE ION, ... | | Authors: | Dienemann, C, Boggild, A, Winther, K.S, Gerdes, K, Brodersen, D.E. | | Deposit date: | 2011-09-01 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the VapBC Toxin-Antitoxin Complex from Shigella flexneri Reveals a Hetero-Octameric DNA-Binding Assembly.
J.Mol.Biol., 414, 2011
|
|
6GYM
 
 | | Structure of a yeast closed complex with distorted DNA (CCdist) | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Dienemann, C, Schwalb, B, Schilbach, S, Cramer, P. | | Deposit date: | 2018-06-30 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Promoter Distortion and Opening in the RNA Polymerase II Cleft.
Mol. Cell, 73, 2019
|
|
6GYK
 
 | | Structure of a yeast closed complex (core CC1) | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Dienemann, C, Schwalb, B, Schilbach, S, Cramer, P. | | Deposit date: | 2018-06-30 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Promoter Distortion and Opening in the RNA Polymerase II Cleft.
Mol. Cell, 73, 2019
|
|
6GYL
 
 | | Structure of a yeast closed complex with distorted DNA (core CCdist) | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Dienemann, C, Schwalb, B, Schilbach, S, Cramer, P. | | Deposit date: | 2018-06-30 | | Release date: | 2018-12-05 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Promoter Distortion and Opening in the RNA Polymerase II Cleft.
Mol. Cell, 73, 2019
|
|
6YYT
 
 | | Structure of replicating SARS-CoV-2 polymerase | | Descriptor: | RNA product, ZINC ION, nsp12, ... | | Authors: | Hillen, H.S, Kokic, G, Farnung, L, Dienemann, C, Tegunov, D, Cramer, P. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of replicating SARS-CoV-2 polymerase.
Nature, 584, 2020
|
|
5IP9
 
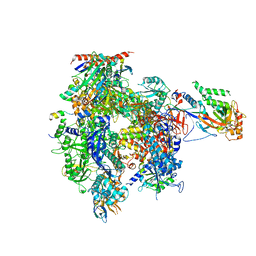 | | Structure of RNA Polymerase II-TFIIF complex | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Transcription initiation complex structures elucidate DNA opening.
Nature, 533, 2016
|
|
5IP7
 
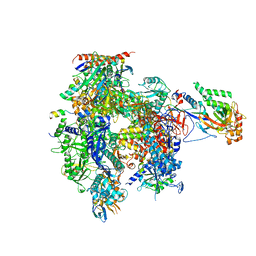 | | Structure of RNA Polymerase II-Tfg1 peptide complex | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Transcription initiation complex structures elucidate DNA opening.
Nature, 533, 2016
|
|
5FZ5
 
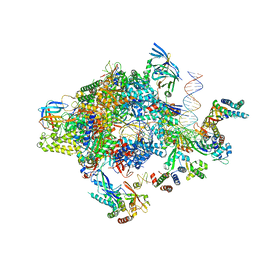 | | Transcription initiation complex structures elucidate DNA opening (CC) | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Transcription Initiation Complex Structures Elucidate DNA Opening
Nature, 533, 2016
|
|
5FYW
 
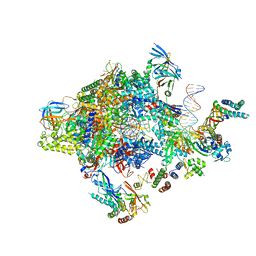 | | Transcription initiation complex structures elucidate DNA opening (OC) | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Transcription Initiation Complex Structures Elucidate DNA Opening
Nature, 533, 2016
|
|
5OQJ
 
 | | STRUCTURE OF YEAST TRANSCRIPTION PRE-INITIATION COMPLEX WITH TFIIH | | Descriptor: | DNA repair helicase RAD25, DNA repair helicase RAD3, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Hantsche, M, Tegunov, D, Dienemann, C, Wigge, C, Henning, U, Cramer, P. | | Deposit date: | 2017-08-11 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structures of transcription pre-initiation complex with TFIIH and Mediator.
Nature, 551, 2017
|
|
5OQM
 
 | | STRUCTURE OF YEAST TRANSCRIPTION PRE-INITIATION COMPLEX WITH TFIIH AND CORE MEDIATOR | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Schilbach, S, Hantsche, M, Tegunov, D, Dienemann, C, Wigge, C, Urlaub, H, Cramer, P. | | Deposit date: | 2017-08-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of transcription pre-initiation complex with TFIIH and Mediator.
Nature, 551, 2017
|
|
6RIE
 
 | | Structure of Vaccinia Virus DNA-dependent RNA polymerase co-transcriptional capping complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Hillen, H.S, Bartuli, J, Grimm, C, Dienemann, C, Bedenk, K, Szalar, A, Fischer, U, Cramer, P. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
6RID
 
 | | Structure of Vaccinia Virus DNA-dependent RNA polymerase elongation complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Hillen, H.S, Bartuli, J, Grimm, C, Dienemann, C, Bedenk, K, Szalar, A, Fischer, U, Cramer, P. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
7B3D
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with AMP at position -4 (structure 3) | | Descriptor: | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'), RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'), SARS-CoV-2 RNA-dependent RNA polymerase nsp12, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
7B3C
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with Remdesivir at position -4 (structure 2) | | Descriptor: | DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*A)-D(P*(RMP))-R(P*GP*UP*G)-3'), Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
7B3B
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with Remdesivir at position -3 (structure 1) | | Descriptor: | DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*G)-D(P*(RMP))-R(P*UP*G)-3'), Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
7OZV
 
 | | SARS-CoV-2 RdRp with Molnupiravir/ NHC in the template strand base-paired with G | | Descriptor: | Non-structural protein 7, Non-structural protein 8, Product RNA, ... | | Authors: | Kabinger, F, Stiller, C, Schmitzova, J, Dienemann, C, Kokic, G, Hillen, H.S, Hoebartner, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OZU
 
 | | SARS-CoV-2 RdRp with Molnupiravir/ NHC in the template strand base-paired with A | | Descriptor: | Non-structural protein 7, Non-structural protein 8, Product RNA, ... | | Authors: | Kabinger, F, Stiller, C, Schmitzova, J, Dienemann, C, Kokic, G, Hillen, H.S, Hoebartner, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ZXE
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZX8
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZWD
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U5 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZX7
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7B7U
 
 | | Cryo-EM structure of mammalian RNA polymerase II in complex with human RPAP2 | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Fianu, I, Dienemann, C, Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of mammalian RNA polymerase II in complex with human RPAP2.
Commun Biol, 4, 2021
|
|
8Q0N
 
 | | HACE1 in complex with RAC1 Q61L | | Descriptor: | E3 ubiquitin-protein ligase HACE1, GUANOSINE-5'-TRIPHOSPHATE, Ras-related C3 botulinum toxin substrate 1, ... | | Authors: | Wolter, M, Duering, J, Dienemann, C, Lorenz, S. | | Deposit date: | 2023-07-28 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PWL
 
 | |
