5I3U
 
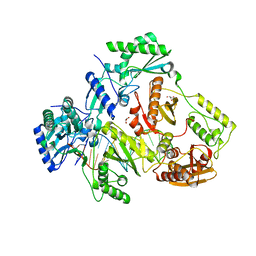 | |
1IHN
 
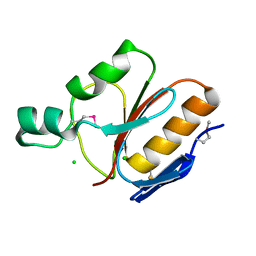 | |
3JYT
 
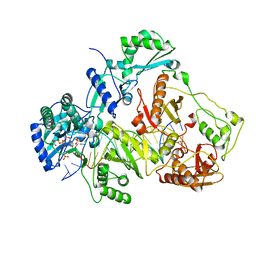 | | K65R mutant HIV-1 reverse transcriptase cross-linked to DS-DNA and complexed with DATP as the incoming nucleotide substrate | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(*A*TP*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | 著者 | Das, K, Arnold, E. | | 登録日 | 2009-09-22 | | 公開日 | 2009-10-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis for the role of the K65r mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance.
J.Biol.Chem., 284, 2009
|
|
3JSM
 
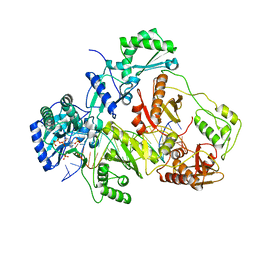 | | K65R mutant HIV-1 reverse transcriptase cross-linked to DS-DNA and complexed with tenofovir-diphosphate as the incoming nucleotide substrate | | 分子名称: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(*A*TP*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), HIV-1 REVERSE TRANSCRIPTASE P51 SUBUNIT, ... | | 著者 | Das, K, Arnold, E. | | 登録日 | 2009-09-10 | | 公開日 | 2009-09-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for the role of the K65r mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance.
J.Biol.Chem., 284, 2009
|
|
1LXY
 
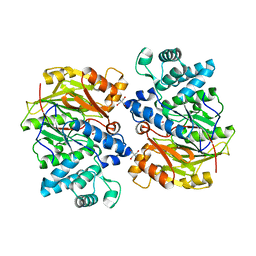 | | Crystal Structure of Arginine Deiminase covalently linked with L-citrulline | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine Deiminase, CITRULLINE | | 著者 | Das, K, Buttler, G.H, Kwiatkowski, V, Yadav, P, Arnold, E. | | 登録日 | 2002-06-06 | | 公開日 | 2004-01-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of arginine deiminase with covalent reaction intermediates; implications for catalytic mechanism
Structure, 12, 2004
|
|
1M0S
 
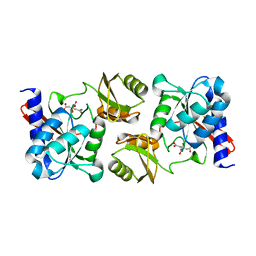 | | NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG ID IR21) | | 分子名称: | CITRIC ACID, Ribose-5-Phosphate Isomerase A | | 著者 | Das, K, Xiao, R, Acton, T, Montelione, G, Arnold, E, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2002-06-14 | | 公開日 | 2002-09-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | D-RIBOSE-5-PHOSPHATE ISOMERASE, IR21
TO BE PUBLISHED
|
|
1P91
 
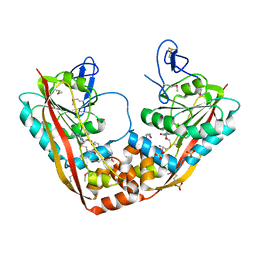 | | Crystal Structure of RlmA(I) enzyme: 23S rRNA n1-G745 methyltransferase (NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER19) | | 分子名称: | Ribosomal RNA large subunit methyltransferase A, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | 著者 | Das, K, Acton, T, Montelione, G, Arnold, E, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-05-08 | | 公開日 | 2004-03-30 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of RlmAI: implications for understanding the 23S rRNA G745/G748-methylation at the macrolide antibiotic-binding site.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1S9R
 
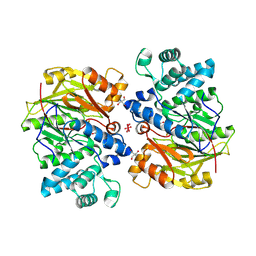 | | CRYSTAL STRUCTURE OF ARGININE DEIMINASE COVALENTLY LINKED WITH A REACTION INTERMEDIATE | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ARGININE, Arginine deiminase, ... | | 著者 | Das, K, Buttler, G.H, Kwiatkowski, V, Clark Jr, A.D, Yadav, P, Arnold, E. | | 登録日 | 2004-02-05 | | 公開日 | 2004-04-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structures of Arginine Deiminase with Covalent Reaction
Intermediates: Implications for Catalytic Mechanism
Structure, 12, 2004
|
|
2RHK
 
 | | Crystal structure of influenza A NS1A protein in complex with F2F3 fragment of human cellular factor CPSF30, Northeast Structural Genomics Targets OR8C and HR6309A | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cleavage and polyadenylation specificity factor subunit 4, NITRATE ION, ... | | 著者 | Das, K, Ma, L.-C, Xiao, R, Radvansky, B, Aramini, J, Zhao, L, Arnold, E, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-10-09 | | 公開日 | 2008-07-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for suppression of a host antiviral response by influenza A virus.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2F6K
 
 | | Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LpR24 | | 分子名称: | MANGANESE (II) ION, metal-dependent hydrolase | | 著者 | Das, K, Xiao, R, Acton, T, Ma, L, Arnold, E, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2005-11-29 | | 公開日 | 2006-01-10 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of ACMDS from Lactobacillus plantarum
To be Published
|
|
3QO9
 
 | | Crystal structure of HIV-1 Reverse Transcriptase (RT) in complex with TSAO-T, a non-nucleoside RT inhibitor (NNRTI) | | 分子名称: | 1-[(5R,6R,8R,9R)-4-amino-9-{[tert-butyl(dimethyl)silyl]oxy}-6-({[tert-butyl(dimethyl)silyl]oxy}methyl)-2,2-dioxido-1,7-dioxa-2-thiaspiro[4.4]non-3-en-8-yl]-5-methylpyrimidine-2,4(1H,3H)-dione, Reverse transcriptase/ ribonuclease H, p51 RT | | 著者 | Das, K, Arnold, E. | | 登録日 | 2011-02-09 | | 公開日 | 2011-05-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of tert-Butyldimethylsilyl-spiroaminooxathioledioxide-thymine (TSAO-T) in Complex with HIV-1 Reverse Transcriptase (RT) Redefines the Elastic Limits of the Non-nucleoside Inhibitor-Binding Pocket.
J.Med.Chem., 54, 2011
|
|
5HRO
 
 | |
5HP1
 
 | |
5I42
 
 | | Structure of HIV-1 Reverse Transcriptase in complex with a DNA aptamer, AZTTP, and CA(2+) ion | | 分子名称: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (38-MER), ... | | 著者 | Das, K, Arnold, E. | | 登録日 | 2016-02-11 | | 公開日 | 2016-06-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Conformational States of HIV-1 Reverse Transcriptase for Nucleotide Incorporation vs Pyrophosphorolysis-Binding of Foscarnet.
Acs Chem.Biol., 11, 2016
|
|
6HAK
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with a double stranded RNA represents the RT transcription initiation complex prior to nucleotide incorporation | | 分子名称: | Gag-Pol polyprotein, MAGNESIUM ION, RNA (5'-R(P*AP*GP*UP*GP*GP*CP*GP*GP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3'), ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2018-08-07 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.95 Å) | | 主引用文献 | Structure of HIV-1 RT/dsRNA initiation complex prior to nucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2BAN
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with JANSSEN-R157208 | | 分子名称: | 5-ETHYL-3-[(2-METHOXYETHYL)METHYLAMINO]-6-METHYL-4-(3-METHYLBENZYL)PYRIDIN-2(1H)-ONE, MANGANESE (II) ION, Reverse transcriptase P51 subunit, ... | | 著者 | Das, K, Arnold, E. | | 登録日 | 2005-10-14 | | 公開日 | 2005-12-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Crystal Structures for HIV-1 Reverse Transcriptase in Complexes with Three Pyridinone Derivatives: A New Class of Non-Nucleoside Inhibitors Effective against a Broad Range of Drug-Resistant Strains.
J.Med.Chem., 48, 2005
|
|
1HNV
 
 | | STRUCTURE OF HIV-1 RT(SLASH)TIBO R 86183 COMPLEX REVEALS SIMILARITY IN THE BINDING OF DIVERSE NONNUCLEOSIDE INHIBITORS | | 分子名称: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | 著者 | Das, K, Ding, J, Arnold, E. | | 登録日 | 1995-03-30 | | 公開日 | 1995-07-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
4R5P
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and a nucleoside triphosphate mimic alpha-carboxy nucleoside phosphonate inhibitor | | 分子名称: | 5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3', 5'-D(*TP*GP*GP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3', HIV-1 reverse transcriptase, ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2014-08-21 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.894 Å) | | 主引用文献 | Alpha-carboxy nucleoside phosphonates as universal nucleoside triphosphate mimics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6ELI
 
 | | Structure of HIV-1 reverse transcriptase (RT) in complex with rilpivirine and an RNase H inhibitor XZ462 | | 分子名称: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Gag-Pol polyprotein, ... | | 著者 | Das, K, Arnold, E. | | 登録日 | 2017-09-29 | | 公開日 | 2018-04-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Developing and Evaluating Inhibitors against the RNase H Active Site of HIV-1 Reverse Transcriptase.
J. Virol., 92, 2018
|
|
6YMV
 
 | |
6YMW
 
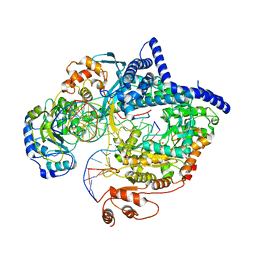 | |
3DXJ
 
 | | Crystal structure of thermus thermophilus rna polymerase holoenzyme in complex with the antibiotic myxopyronin | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, BACTERIAL RNA POLYMERASE BETA SUBUNIT; CHAIN C, M, ... | | 著者 | Das, K, Arnold, E. | | 登録日 | 2008-07-24 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The RNA polymerase "switch region" is a target for inhibitors.
Cell(Cambridge,Mass.), 135, 2008
|
|
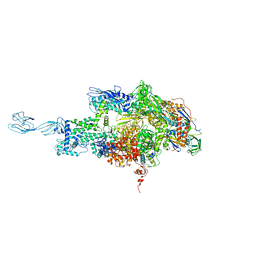
5HLF
 
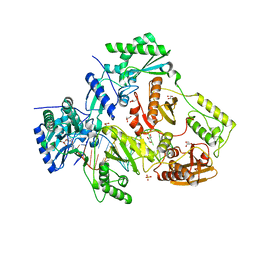 | |
6FBV
 
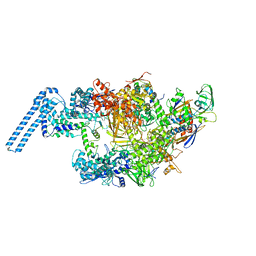 | | Single particle cryo em structure of Mycobacterium tuberculosis RNA polymerase in complex with Fidaxomicin | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Das, K, Lin, W, Ebright, E. | | 登録日 | 2017-12-19 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
3S24
 
 | | Crystal structure of human mRNA guanylyltransferase | | 分子名称: | SULFATE ION, mRNA-capping enzyme | | 著者 | Das, K, Chu, C, Thyminski, J.R, Bauman, J.D, Guan, R, Qiu, W, Montelione, G.T, Arnold, E, Shatkin, A.J. | | 登録日 | 2011-05-16 | | 公開日 | 2011-06-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.0137 Å) | | 主引用文献 | Structure of the guanylyltransferase domain of human mRNA capping enzyme.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
