2M8D
 
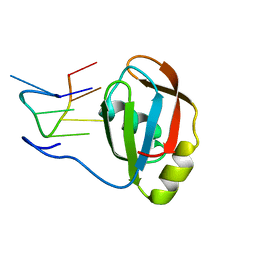 | | Structure of SRSF1 RRM2 in complex with the RNA 5'-UGAAGGAC-3' | | Descriptor: | RNA (5'-R(*UP*GP*AP*AP*GP*GP*AP*C)-3'), Serine/arginine-rich splicing factor 1 | | Authors: | Clery, A, Sinha, R, Anczukow, O, Corrionero, A, Moursy, A, Daubner, G, Valcarcel, J, Krainer, A.R, Allain, F.H.T. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2KXN
 
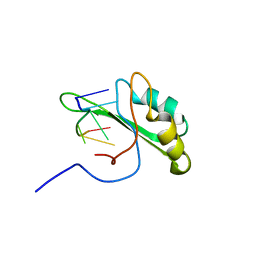 | | NMR structure of human Tra2beta1 RRM in complex with AAGAAC RNA | | Descriptor: | 5'-R(*AP*AP*GP*AP*AP*C)-3', Transformer-2 protein homolog beta | | Authors: | Clery, A, Jayne, S, Benderska, N, Dominguez, C, Stamm, S, Allain, F.H.-T. | | Deposit date: | 2010-05-10 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1
Nat.Struct.Mol.Biol., 18, 2011
|
|
6HPJ
 
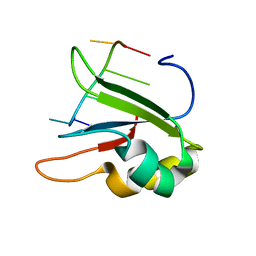 | | Structure of human SRSF1 RRM1 bound to AACAAA RNA | | Descriptor: | Immunoglobulin G-binding protein G,Serine/arginine-rich splicing factor 1, RNA (5'-R(*AP*AP*CP*AP*AP*A)-3') | | Authors: | Allain, F.T.H, Clery, A. | | Deposit date: | 2018-09-21 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of SRSF1 RRM1 bound to RNA reveals an unexpected bimodal mode of interaction and explains its involvement in SMN1 exon7 splicing.
Nat Commun, 12, 2021
|
|
7QDD
 
 | |
7QDE
 
 | |
5OR0
 
 | |
2MB0
 
 | |
2LEA
 
 | | Solution structure of human SRSF2 (SC35) RRM | | Descriptor: | Serine/arginine-rich splicing factor 2 | | Authors: | Daubner, G.M, Clery, A, Jayne, S, Stevenin, J, Allain, F.H.-T. | | Deposit date: | 2011-06-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well.
Embo J., 31, 2012
|
|
2LEC
 
 | | Solution structure of human SRSF2 (SC35) RRM in complex with 5'-UGGAGU-3' | | Descriptor: | RNA (5'-R(*UP*GP*GP*AP*GP*U)-3'), Serine/arginine-rich splicing factor 2 | | Authors: | Daubner, G.M, Clery, A, Jayne, S, Stevenin, J, Allain, F.H.-T. | | Deposit date: | 2011-06-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well.
Embo J., 31, 2012
|
|
2LEB
 
 | | Solution structure of human SRSF2 (SC35) RRM in complex with 5'-UCCAGU-3' | | Descriptor: | RNA (5'-R(*UP*CP*CP*AP*GP*U)-3'), Serine/arginine-rich splicing factor 2 | | Authors: | Daubner, G.M, Clery, A, Jayne, S, Stevenin, J, Allain, F.H.-T. | | Deposit date: | 2011-06-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well.
Embo J., 31, 2012
|
|
7AEP
 
 | | Solution structure of U1-A RRM2 (190-282) | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Campagne, S, Allain, F.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | An in vitro reconstituted U1 snRNP allows the study of the disordered regions of the particle and the interactions with proteins and ligands.
Nucleic Acids Res., 49, 2021
|
|
6HMI
 
 | |
5MPL
 
 | | hnRNP A1 RRM2 in complex with 5'-UCAGUU-3' RNA | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, RNA UCAGUU | | Authors: | Barraud, P, Allain, F.H.-T. | | Deposit date: | 2016-12-16 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Tandem hnRNP A1 RNA recognition motifs act in concert to repress the splicing of survival motor neuron exon 7.
Elife, 6, 2017
|
|
5MPG
 
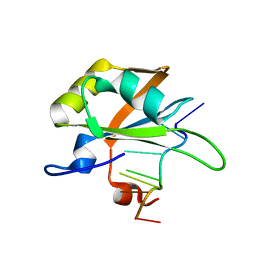 | |
6HMO
 
 | |
6GBM
 
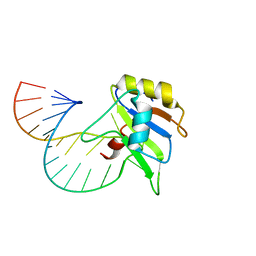 | | Solution structure of FUS-RRM bound to stem-loop RNA | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*GP*AP*UP*UP*AP*CP*AP*AP*UP*UP*CP*UP*AP*UP*UP*UP*GP*CP*C)-3'), RNA-binding protein FUS | | Authors: | Loughlin, F.E, Allain, F.H.-T. | | Deposit date: | 2018-04-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Mol. Cell, 73, 2019
|
|
6G99
 
 | | Solution structure of FUS-ZnF bound to UGGUG | | Descriptor: | RNA (5'-R(*UP*GP*GP*UP*G)-3'), RNA-binding protein FUS, ZINC ION | | Authors: | Loughlin, F.E, Allain, F.H.-T. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Mol. Cell, 73, 2019
|
|
7ZO1
 
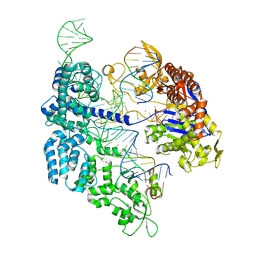 | | SpCas9 bound to CD34 off-target9 DNA substrate | | Descriptor: | CD34 off-target9 DNA non-target strand, CD34 off-target9 DNA target strand, CD34 sgRNA, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-04-23 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7ZEV
 
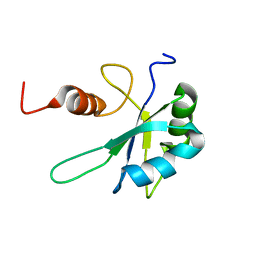 | | Free form of extended Cyp33-RRM | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEW
 
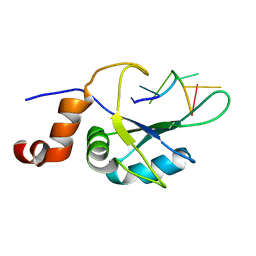 | | Complex Cyp33-RRM : AAUAAA RNA | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E, RNA (5'-R(*AP*AP*UP*AP*AP*A)-3') | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEZ
 
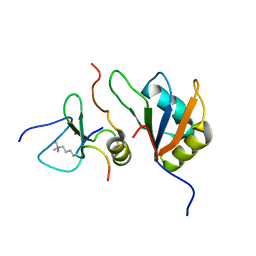 | | Trimolecular complex Cyp33-RRMdelta alpha : MLL1-PHD3 : H3K4me3 | | Descriptor: | Histone H3, Isoform 3 of Peptidyl-prolyl cis-trans isomerase E, MLL cleavage product N320, ... | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-05-03 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEX
 
 | | Complex Cyp33-RRMdelta alpha : UAAUGUCG RNA | | Descriptor: | Isoform 3 of Peptidyl-prolyl cis-trans isomerase E, RNA (5'-R(*UP*AP*AP*UP*GP*UP*CP*G)-3') | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEY
 
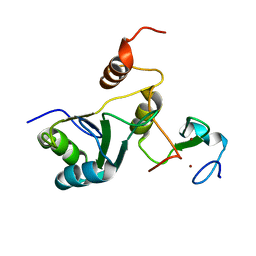 | | Complex Cyp33-RRM : MLL1-PHD3 | | Descriptor: | MLL cleavage product N320, Peptidyl-prolyl cis-trans isomerase E, ZINC ION | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7QQR
 
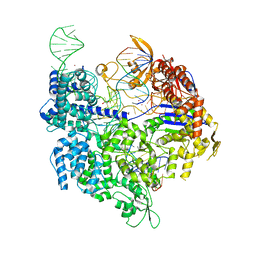 | | SpCas9 bound to AAVS1 off-target5 DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, AAVS1 off-target5 non-target strand, AAVS1 off-target5 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQV
 
 | | SpCas9 bound to FANCF off-target3 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target3 non-target strand, FANCF off-target3 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
