5CS5
 
 | | The structure of the NK1 fragment of HGF/SF complexed with PIPES | | Descriptor: | Hepatocyte growth factor, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID) | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CT2
 
 | | The structure of the NK1 fragment of HGF/SF complexed with CAPS | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CT3
 
 | | The structure of the NK1 fragment of HGF/SF complexed with 2FA | | Descriptor: | 3-hydroxypropane-1-sulfonic acid, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CS3
 
 | | The structure of the NK1 fragment of HGF/SF complexed with (H)EPPS | | Descriptor: | 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
7A2I
 
 | |
7A7C
 
 | |
7A7A
 
 | |
7A8Z
 
 | |
7AA3
 
 | |
7AG8
 
 | |
7OPL
 
 | |
7OTP
 
 | | DNA-PKcs in complex with ATPgammaS-Mg | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTY
 
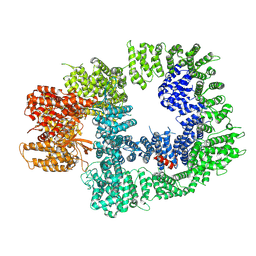 | | DNA-PKcs in complex with M3814 | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTM
 
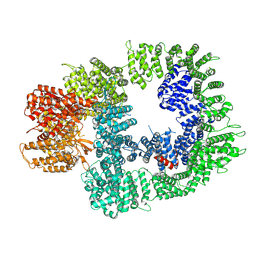 | | Cryo-EM structure of DNA-PKcs in complex with NU7441 | | Descriptor: | 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTW
 
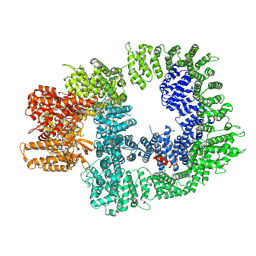 | | DNA-PKcs in complex with AZD7648 | | Descriptor: | 7-methyl-2-[(7-methyl-[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(oxan-4-yl)purin-8-one, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTV
 
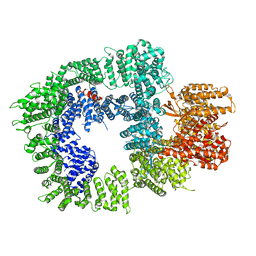 | | DNA-PKcs in complex with wortmannin | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7PBD
 
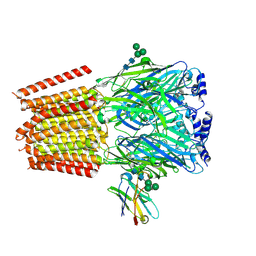 | | a1b3 GABA-A receptor + GABA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Miller, P.S, Kasaragod, V.B. | | Deposit date: | 2021-08-02 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Mechanisms of inhibition and activation of extrasynaptic alpha beta GABA A receptors.
Nature, 602, 2022
|
|
7PC0
 
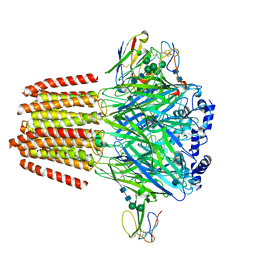 | | GABA-A receptor bound by a-Cobratoxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-cobratoxin, ... | | Authors: | Kasaragod, V.B, Miller, P.S. | | Deposit date: | 2021-08-03 | | Release date: | 2022-02-09 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanisms of inhibition and activation of extrasynaptic alpha beta GABA A receptors.
Nature, 602, 2022
|
|
7PBZ
 
 | | a1b3 GABA-A receptor + GABA + Zn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DECANE, ... | | Authors: | Miller, P.S, Kasaragod, V.B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Mechanisms of inhibition and activation of extrasynaptic alpha beta GABA A receptors.
Nature, 602, 2022
|
|
7Z6O
 
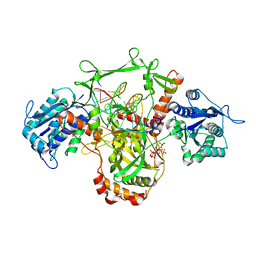 | | X-Ray studies of Ku70/80 reveal the binding site for IP6 | | Descriptor: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Varela, P.F, Charbonnier, J.B. | | Deposit date: | 2022-03-14 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
7ZT6
 
 | |
8ASC
 
 | | Ku70/80 binds to the Ku-binding motif of PAXX | | Descriptor: | DNA (5'-D(P*CP*GP*GP*AP*TP*CP*GP*AP*GP*GP*GP*CP*CP*CP*GP*AP*TP*AP*T)-3'), DNA (5'-D(P*GP*GP*GP*CP*CP*CP*TP*CP*GP*AP*TP*CP*CP*G)-3'), Protein PAXX, ... | | Authors: | Seif El Dahan, M, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
8BOT
 
 | |
8BSC
 
 | |
8BR2
 
 | | CryoEM structure of the post-synaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*GP*CP*GP*AP*GP*CP*TP*CP*GP*AP*TP*GP*CP*AP*CP*CP*TP*CP*CP*A)-3'), ... | | Authors: | Appleby, R, Bollschweiler, D, Pellegrini, L. | | Deposit date: | 2022-11-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A metal ion-dependent mechanism of RAD51 nucleoprotein filament disassembly.
Iscience, 26, 2023
|
|
