6LNI
 
 | | Cryo-EM structure of amyloid fibril formed by full-length human prion protein | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Wang, Q, Guan, Z.Y, Tao, J, Li, X.N, Hao, M.M, Chen, J, Zhang, D.L, Zhu, H.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2019-12-30 | | Release date: | 2020-06-10 | | Last modified: | 2020-06-24 | | Method: | ELECTRON MICROSCOPY (2.702 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human prion protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7CJL
 
 | | Metallo-Beta-Lactamase VIM-2 in complex with (S)-N-(3-(2H-tetrazol-5-yl)phenyl)-3-mercapto-2-methylpropanamide | | Descriptor: | (S)-N-(3-(2H-tetrazol-5-yl)phenyl)-3-mercapto-2-methylpropanamide, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Yan, Y.-H, Chen, J, Zhan, Z, Yu, Z.-J, Li, G, Li, G.-B, Guo, L, Wu, Y. | | Deposit date: | 2020-07-11 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Discovery of mercaptopropanamide-substituted aryl tetrazoles as new broad-spectrum metallo-beta-lactamase inhibitors.
Rsc Adv, 10, 2020
|
|
7DWV
 
 | | Cryo-EM structure of amyloid fibril formed by familial prion disease-related mutation E196K | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Li, X.N, Dang, H.B, Ma, Y.Y, Wang, Q, Wang, C, Sun, Y.P, Chen, J, Li, D, Zhang, D.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-01-18 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
5ZN8
 
 | | Crystal structure of nicotinamidase PncA from Bacillus subtilis | | Descriptor: | Isochorismatase, ZINC ION | | Authors: | Shang, F, Chen, J, Wang, L, Xu, Y. | | Deposit date: | 2018-04-08 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the nicotinamidase/pyrazinamidase PncA from Bacillus subtilis.
Biochem.Biophys.Res.Commun., 503, 2018
|
|
6A8L
 
 | | Crystal structure of nicotinamidase/ pyrazinamidase PncA from Bacillus subtilis | | Descriptor: | Isochorismatase, ZINC ION | | Authors: | Shang, F, Chen, J, Wang, L, Xu, Y. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the nicotinamidase/pyrazinamidase PncA from Bacillus subtilis.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
6BZO
 
 | | Mtb RNAP Holo/RbpA/Fidaxomicin/upstream fork DNA | | Descriptor: | DNA (26-MER), DNA (32-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2017-12-25 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
6BHU
 
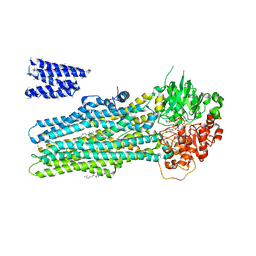 | |
6C0V
 
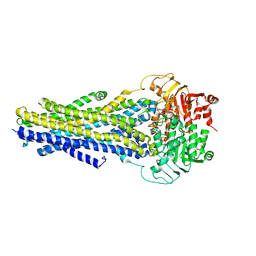 | |
6C3P
 
 | | Cryo-EM structure of human KATP bound to ATP and ADP in propeller form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Lee, K.P.K, Chen, J, MacKinnon, R. | | Deposit date: | 2018-01-10 | | Release date: | 2018-01-24 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Molecular structure of human KATP in complex with ATP and ADP.
Elife, 6, 2017
|
|
6C3O
 
 | | Cryo-EM structure of human KATP bound to ATP and ADP in quatrefoil form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Lee, K.P.K, Chen, J, MacKinnon, R. | | Deposit date: | 2018-01-10 | | Release date: | 2018-01-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of human KATP in complex with ATP and ADP
Elife, 6, 2017
|
|
7YDL
 
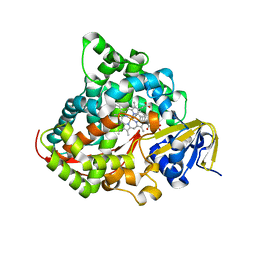 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268I/A184V/A82T in complex with N-imidazolyl-hexanoyl-L-phenylalanine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A/T268I/A184V/A82T in complex with N-imidazolyl-hexanoyl-L-phenylalanine
To Be Published
|
|
6M7J
 
 | | Mycobacterium tuberculosis RNAP with RbpA/us fork and Corallopyronin | | Descriptor: | DNA (26-MER), DNA (31-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
6MSM
 
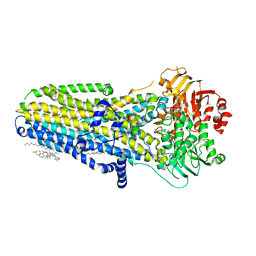 | | Phosphorylated, ATP-bound human cystic fibrosis transmembrane conductance regulator (CFTR) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, ... | | Authors: | Zhang, Z, Liu, F, Chen, J. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular structure of the ATP-bound, phosphorylated human CFTR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2OJU
 
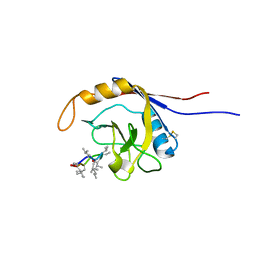 | | X-ray structure of complex of human cyclophilin J with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE-LIKE 3 | | Authors: | Xia, Z, Huang, L. | | Deposit date: | 2007-01-14 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Cyclophilin J, a Novel Peptidyl-Prolyl Isomerase, Can Induce Cellular G1/S Arrest and Repress the Growth of Hepatocellular Carcinoma
To be Published
|
|
2OK3
 
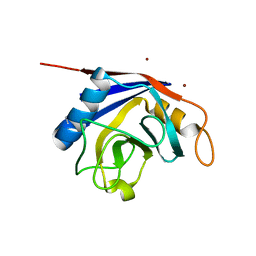 | | X-ray structure of human cyclophilin J at 2.0 angstrom | | Descriptor: | NICKEL (II) ION, Peptidyl-prolyl cis-trans isomerase-like 3 | | Authors: | Xia, Z. | | Deposit date: | 2007-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Cyclophilin J, a novel peptidyl-prolyl isomerase, can induce cellular G1/S arrest and repress the growth of Hepatocellular carcinoma
To be Published
|
|
2O7Y
 
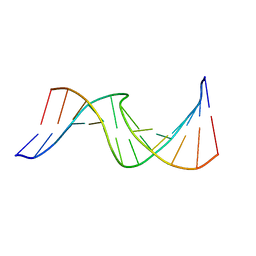 | | Duplex DNA containing an abasic site with an opposite T (alpha anomer) in 5'-G_AC-3' (10 structure ensemble and averaged structure) | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(ORP)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*TP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-12-11 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | DNA oligonucleotides with A, T, G or C opposite an abasic site: structure and dynamics.
Nucleic Acids Res., 36, 2008
|
|
3ED0
 
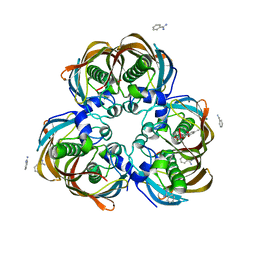 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with emodin | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, BENZAMIDINE, ... | | Authors: | Zhang, L, Zhang, H, Liu, W, Guo, Y, Shen, X, Jiang, H. | | Deposit date: | 2008-09-02 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Emodin targets the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: enzymatic inhibition assay with crystal structural and thermodynamic characterization
BMC MICROBIOL., 9, 2009
|
|
7WZ5
 
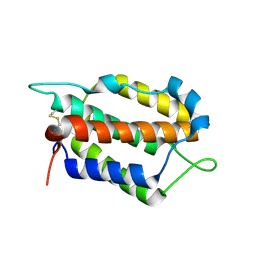 | | Larimichthys crocea IFNi | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon C | | Authors: | Chen, J.J. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Molecular and Structural Basis of Receptor Binding and Signaling of a Fish Type I IFN with Three Disulfide Bonds.
J Immunol., 209, 2022
|
|
3CYY
 
 | |
7M10
 
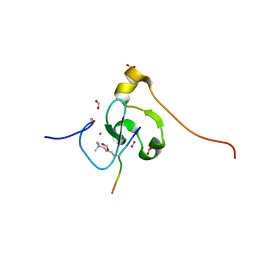 | | PHF2 PHD Domain Complexed with Peptide From N-terminus of VRK1 | | Descriptor: | FORMIC ACID, Lysine-specific demethylase PHF2, Serine/threonine-protein kinase VRK1 N-terminus peptide, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2021-03-11 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Histone H3 N-terminal mimicry drives a novel network of methyl-effector interactions.
Biochem.J., 478, 2021
|
|
7DR6
 
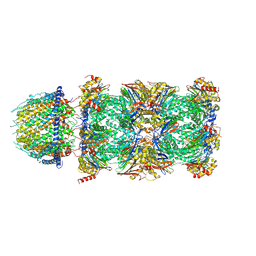 | | PA28alpha-beta in complex with immunoproteasome | | Descriptor: | Proteasome activator complex subunit 1, Proteasome activator complex subunit 2, Proteasome subunit alpha type-1, ... | | Authors: | Cong, Y, Xu, C. | | Deposit date: | 2020-12-26 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM of mammalian PA28 alpha beta-iCP immunoproteasome reveals a distinct mechanism of proteasome activation by PA28 alpha beta.
Nat Commun, 12, 2021
|
|
7DR7
 
 | | bovine 20S immunoproteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2020-12-26 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM of mammalian PA28 alpha beta-iCP immunoproteasome reveals a distinct mechanism of proteasome activation by PA28 alpha beta.
Nat Commun, 12, 2021
|
|
7DRW
 
 | | Bovine 20S immunoproteasome in complex with two human PA28alpha-beta activators | | Descriptor: | Proteasome activator complex subunit 1, Proteasome activator complex subunit 2, Proteasome subunit alpha type-1, ... | | Authors: | Cong, Y, Xu, C. | | Deposit date: | 2020-12-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of mammalian PA28 alpha beta-iCP immunoproteasome reveals a distinct mechanism of proteasome activation by PA28 alpha beta.
Nat Commun, 12, 2021
|
|
6IIJ
 
 | | Cryo-EM structure of CV-A10 mature virion | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Chen, J.H, Ye, X.H, Cong, Y, Huang, Z. | | Deposit date: | 2018-10-06 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Coxsackievirus A10 atomic structure facilitating the discovery of a broad-spectrum inhibitor against human enteroviruses.
Cell Discov, 5, 2019
|
|
6IIO
 
 | | Cryo-EM structure of CV-A10 native empty particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Chen, J.H, Ye, X.H, Cong, Y, Huang, Z. | | Deposit date: | 2018-10-07 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Coxsackievirus A10 atomic structure facilitating the discovery of a broad-spectrum inhibitor against human enteroviruses.
Cell Discov, 5, 2019
|
|
