7YJ4
 
 | | Cryo-EM structure of the INSL5-bound human relaxin family peptidereceptor 4 (RXFP4)-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-07-19 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7YK7
 
 | | Cryo-EM structure of the DC591053-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Xu, H.E, Yang, D.H, Liu, H, Wang, M.W. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7YK6
 
 | | Cryo-EM structure of the compound 4-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | Descriptor: | 1-[2-(4-chlorophenyl)ethyl]-3-[(7-ethyl-5-oxidanyl-1H-indol-3-yl)methylideneamino]guanidine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7WBJ
 
 | | Cryo-EM structure of N-terminal modified human vasoactive intestinal polypeptide receptor 2 (VIP2R) in complex with PACAP27 and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, Y.N, Feng, W.B, Zhou, Q.T, Liang, A.Y, Li, J, Dai, A.T, Zhao, F.H, Yan, J.H, Chen, C.W, Li, H, Zhao, L.H, Xia, T, Jiang, Y, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-12-16 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | A distinctive ligand recognition mechanism by the human vasoactive intestinal polypeptide receptor 2.
Nat Commun, 13, 2022
|
|
7VQX
 
 | | Cryo-EM structure of human vasoactive intestinal polypeptide receptor 2 (VIP2R) in complex with PACAP27 and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, Y.N, Feng, W.B, Zhou, Q.T, Liang, A.Y, Li, J, Dai, A.T, Zhao, F.H, Yan, J.H, Chen, C.W, Li, H, Zhao, L.H, Xia, T, Jiang, Y, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-10-21 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | A distinctive ligand recognition mechanism by the human vasoactive intestinal polypeptide receptor 2.
Nat Commun, 13, 2022
|
|
8FC1
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and erythromycin at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8FC2
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and azithromycin at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8FC6
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and telithromycin at 2.35A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-26 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8FC5
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and azithromycin at 2.65A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-26 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8FC4
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and erythromycin at 2.45A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-26 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8FC3
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and telithromycin at 2.60A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
7MD7
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with triphenylphosphonium analog of chloramphenicol CAM-C4-TPP and protein Y (YfiA) at 2.80A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Pavlova, J.A, Lukianov, D.A, Tereshchenkov, A.G, Makarov, G.I, Khairullina, Z.Z, Tashlitsky, V.N, Paleskava, A, Konevega, A.L, Bogdanov, A.A, Osterman, I.A, Sumbatyan, N.V, Polikanov, Y.S. | | Deposit date: | 2021-04-03 | | Release date: | 2021-04-21 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and Action of Triphenylphosphonium Analog of Chloramphenicol upon the Bacterial Ribosome.
Antibiotics, 10, 2021
|
|
7WUH
 
 | | SARS-CoV-2 Spike in complex with Fab of m31A7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2022-02-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models.
Sci Transl Med, 14, 2022
|
|
7WUE
 
 | | Crystal structure of SARS-CoV-2 Receptor Binding Domain in complex with the monoclonal antibody m31A7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mohapatra, A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models.
Sci Transl Med, 14, 2022
|
|
7PAD
 
 | | The crystal structure of DW-0254 in complex with PDE6D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Beaumont, E, Williams, D. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Validation of a small molecule inhibitor of PDE6D-RAS interaction with favorable anti-leukemic effects.
Blood Cancer J, 12, 2022
|
|
7PAC
 
 | | The crystal structure of PDE6D in the apo state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NICKEL (II) ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Beaumont, E, Williams, D. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Validation of a small molecule inhibitor of PDE6D-RAS interaction with favorable anti-leukemic effects.
Blood Cancer J, 12, 2022
|
|
7PAE
 
 | | The crystal structure of Deltarasin in complex with PDE6D | | Descriptor: | ACETIC ACID, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, deltarasin | | Authors: | Beaumont, E, Williams, D. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Validation of a small molecule inhibitor of PDE6D-RAS interaction with favorable anti-leukemic effects.
Blood Cancer J, 12, 2022
|
|
6RSA
 
 | |
8W76
 
 | | Crystal structure of d(CGTATACG)2 duplex | | Descriptor: | DNA (5'-D(P*CP*GP*TP*AP*TP*AP*CP*G)-3'), MANGANESE (II) ION | | Authors: | Satange, R.B, Peng, C.L, Hou, M.H. | | Deposit date: | 2023-08-30 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Targeting DNA junction sites by bis-intercalators induces topological changes with potent antitumor effects.
Nucleic Acids Res., 52, 2024
|
|
8W7W
 
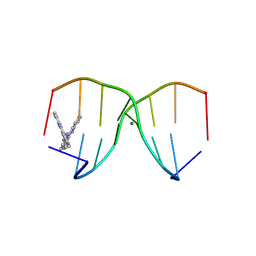 | | Crystal structure of d(CGTATACG)2 with acridine complex | | Descriptor: | DNA (5'-D(P*CP*GP*TP*AP*TP*AP*CP*G)-3'), MAGNESIUM ION, ~{N},~{N}'-di(acridin-9-yl)pentane-1,5-diamine | | Authors: | Huang, S.C, Satange, R.B, Hou, M.H. | | Deposit date: | 2023-08-31 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting DNA junction sites by bis-intercalators induces topological changes with potent antitumor effects.
Nucleic Acids Res., 52, 2024
|
|
8WQ7
 
 | | Crystal structure of d(CGTATACG)2 with a four-carbon linker containing diacridine compound | | Descriptor: | DNA (5'-D(P*CP*GP*TP*AP*TP*AP*CP*G)-3'), MANGANESE (II) ION, N,N'-di(acridin-9-yl)butane-1,4-diamine | | Authors: | Huang, S.C, Satange, R.B, Hou, M.H. | | Deposit date: | 2023-10-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting DNA junction sites by bis-intercalators induces topological changes with potent antitumor effects.
Nucleic Acids Res., 52, 2024
|
|
6ND5
 
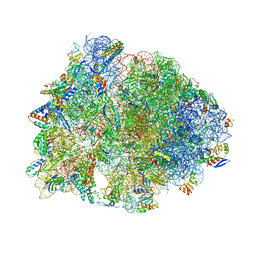 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with chloramphenicol and bound to mRNA and A-, P-, and E-site tRNAs at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-03-20 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
6ND6
 
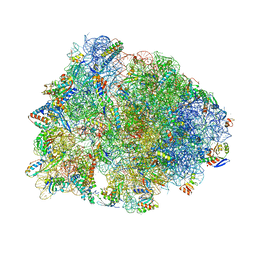 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with erythromycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.85A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-02-20 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
6JXG
 
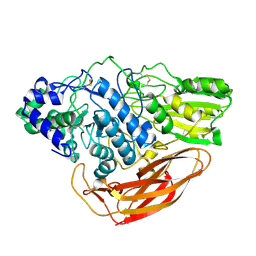 | | Crystasl Structure of Beta-glucosidase D2-BGL from Chaetomella Raphigera | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-glucosidase, alpha-D-mannopyranose | | Authors: | Wang, A.H.-J, Lee, C.C, Kao, M.R, Ho, T.H.D. | | Deposit date: | 2019-04-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chaetomella raphigerabeta-glucosidase D2-BGL has intriguing structural features and a high substrate affinity that renders it an efficient cellulase supplement for lignocellulosic biomass hydrolysis.
Biotechnol Biofuels, 12, 2019
|
|
1RUV
 
 | | RIBONUCLEASE A-URIDINE VANADATE COMPLEX: HIGH RESOLUTION RESOLUTION X-RAY STRUCTURE (1.3 A) | | Descriptor: | RIBONUCLEASE A, TERTIARY-BUTYL ALCOHOL, URIDINE-2',3'-VANADATE | | Authors: | Ladner, J.E, Wladkowski, B, Svensson, L.A, Sjolin, L, Gilliland, G.L. | | Deposit date: | 1995-07-27 | | Release date: | 1997-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | X-ray structure of a ribonuclease A-uridine vanadate complex at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
