7OPB
 
 | | IL7R in complex with an antagonist | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IL7R binder, ... | | Authors: | Markovic, I, Verschueren, K.H.G, Verstraete, K, Savvides, S.N. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
7JZL
 
 | |
7JZN
 
 | |
7JZM
 
 | |
7JZU
 
 | |
7RDH
 
 | |
6MAR
 
 | |
6JFP
 
 | | Crystal structure of the beta-glucosidase Bgl15 | | Descriptor: | beta-D-glucopyranose, beta-glucosidase 15 | | Authors: | Xie, W, Chen, R. | | Deposit date: | 2019-02-11 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineering of beta-Glucosidase Bgl15 with Simultaneously Enhanced Glucose Tolerance and Thermostability To Improve Its Performance in High-Solid Cellulose Hydrolysis.
J.Agric.Food Chem., 68, 2020
|
|
4BW5
 
 | | Crystal structure of human two pore domain potassium ion channel TREK2 (K2P10.1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CADMIUM ION, POTASSIUM CHANNEL SUBFAMILY K MEMBER 10, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Dong, L, Quigley, A, Shrestha, L, Mukhopadhyay, S, Strain-Damerell, C, Goubin, S, Grieben, M, Shintre, C.A, Mackenzie, A, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2013-06-30 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | K2P Channel Gating Mechanisms Revealed by Structures of Trek-2 and a Complex with Prozac
Science, 347, 2015
|
|
5OD4
 
 | |
6DNE
 
 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS660 | | Descriptor: | Bromodomain-containing protein 4, N,N'-[ethane-1,2-diylbis(oxyethane-2,1-diyl)]bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.958 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DJC
 
 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS645 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-(decane-1,10-diyl)bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-05-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4XDK
 
 | | Crystal structure of human two pore domain potassium ion channel TREK2 (K2P10.1) in complex with norfluoxetine | | Descriptor: | (3R)-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine, (3S)-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Mackenzie, A, Mukhopadhyay, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | K2P channel gating mechanisms revealed by structures of TREK-2 and a complex with Prozac.
Science, 347, 2015
|
|
4XDL
 
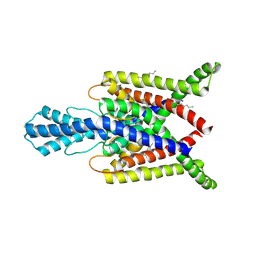 | | Crystal structure of human two pore domain potassium ion channel TREK2 (K2P10.1) in complex with a brominated fluoxetine derivative. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[2-bromanyl-4-(trifluoromethyl)phenoxy]-N-methyl-3-phenyl-propan-1-amine, CADMIUM ION, ... | | Authors: | Mackenzie, A, Pike, A.C.W, Dong, Y.Y, Mukhopadhyay, S, Ruda, G.F, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | K2P channel gating mechanisms revealed by structures of TREK-2 and a complex with Prozac.
Science, 347, 2015
|
|
4XDJ
 
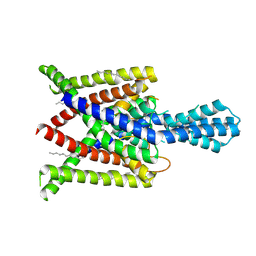 | | Crystal structure of human two pore domain potassium ion channel TREK2 (K2P10.1) in an alternate conformation (FORM 2) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, POTASSIUM CHANNEL SUBFAMILY K MEMBER 10, POTASSIUM ION, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Mackenzie, A, Mukhopadhyay, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | K2P channel gating mechanisms revealed by structures of TREK-2 and a complex with Prozac.
Science, 347, 2015
|
|
6Q1X
 
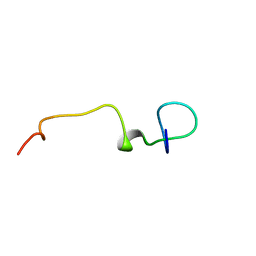 | | Lasso peptide pandonodin | | Descriptor: | Pandonodin | | Authors: | Link, A.J, Cheung-Lee, W.L. | | Deposit date: | 2019-08-06 | | Release date: | 2019-12-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pandonodin: A Proteobacterial Lasso Peptide with an Exceptionally Long C-Terminal Tail.
Acs Chem.Biol., 14, 2019
|
|
5F6L
 
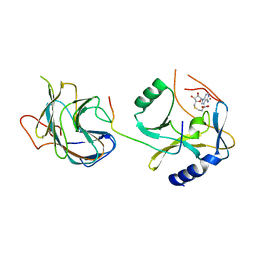 | | The crystal structure of MLL1 (N3861I/Q3867L) in complex with RbBP5 and Ash2L | | Descriptor: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F59
 
 | | The crystal structure of MLL3 SET domain | | Descriptor: | Histone-lysine N-methyltransferase 2C, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-04 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F5E
 
 | |
5F6K
 
 | | Crystal structure of the MLL3-Ash2L-RbBP5 complex | | Descriptor: | Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
6D2P
 
 | |
7KUW
 
 | |
8JAH
 
 | | Crystal structure of human CLEC12A C-type lectin domain | | Descriptor: | C-type lectin domain family 12 member A, POTASSIUM ION, SULFATE ION | | Authors: | Lei, Q, Tang, H, Dong, X. | | Deposit date: | 2023-05-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Mechanistic insights into the C-type lectin receptor CLEC12A-mediated immune recognition of monosodium urate crystal.
J.Biol.Chem., 300, 2024
|
|
5H4B
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5DZN
 
 | | human T-cell immunoglobulin and mucin domain protein 4 | | Descriptor: | T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Yuan, S, Rao, Z, Wang, X. | | Deposit date: | 2015-09-25 | | Release date: | 2015-11-25 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIM-1 acts a dual-attachment receptor for Ebolavirus by interacting directly with viral GP and the PS on the viral envelope.
Protein Cell, 6, 2015
|
|
