7KCR
 
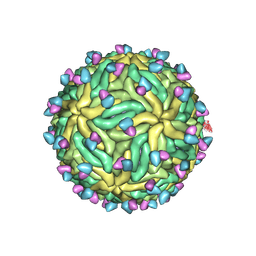 | | Cryo-EM structure of Zika virus in complex with E protein cross-linking human monoclonal antibody ADI30056 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADI30056 Fab heavy chain variable region, ADI30056 Fab light chain variable region, ... | | Authors: | Sevvana, M, Rogers, T.F, Miller, A.S, Long, F, Klose, T, Beutler, N, Lai, Y.C, Parren, M, Walker, L.M, Buda, G, Burton, D.R, Rossmann, M.G, Kuhn, R.J. | | Deposit date: | 2020-10-07 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Zika Virus Specific Neutralization in Subsequent Flavivirus Infections.
Viruses, 12, 2020
|
|
7RXP
 
 | |
7RXI
 
 | |
7RXJ
 
 | |
7S0X
 
 | |
7RXL
 
 | |
7SJS
 
 | |
8SWH
 
 | |
7TP3
 
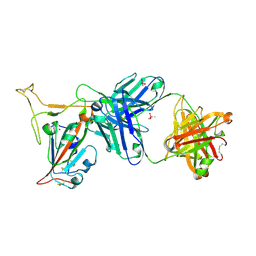 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K288.2 | | Descriptor: | CACODYLATE ION, K288.2 heavy chain, K288.2 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7TP4
 
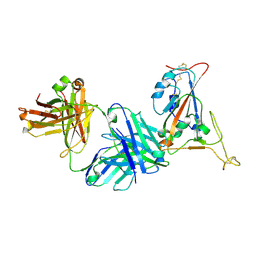 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K398.22 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, K398.22 heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
8EKF
 
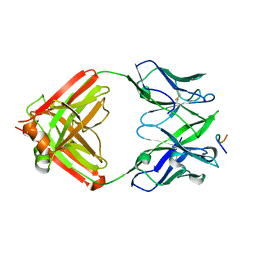 | |
7LQ7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV503 and COVA1-16 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 13, 2021
|
|
8DGX
 
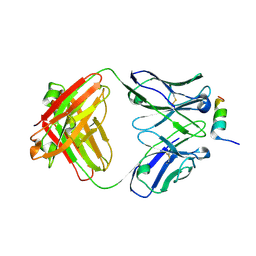 | |
8DGU
 
 | |
8DGV
 
 | |
8DGW
 
 | |
7NAB
 
 | | Crystal structure of human neutralizing mAb CV3-25 binding to SARS-CoV-2 S MPER peptide 1140-1165 | | Descriptor: | CITRIC ACID, CV3-25 Fab Heavy Chain, CV3-25 Fab Light Chain, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-06-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern.
Cell Rep, 38, 2022
|
|
8DYT
 
 | | Cryo-EM structure of 227 Fab in complex with (NPNA)8 peptide | | Descriptor: | 227 Fab heavy chain, 227 Fab light chain, Circumsporozoite protein | | Authors: | Martin, G.M, Ward, A.B. | | Deposit date: | 2022-08-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Affinity-matured homotypic interactions induce spectrum of PfCSP structures that influence protection from malaria infection.
Nat Commun, 14, 2023
|
|
8DYW
 
 | |
8DZ5
 
 | |
8DYX
 
 | |
8DZ4
 
 | |
8DYY
 
 | |
8DZ3
 
 | |
