4WGH
 
 | | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution | | Descriptor: | ACETATE ION, Aldehyde reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bacal, P, Shabalin, I.G, Cooper, D.R, Hillerich, B.S, Zimmerman, M.D, Chowdhury, S, Hammonds, J, Al Obaidi, N, Gizzi, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution
to be published
|
|
4WKC
 
 | | Crystal structure of Escherichia coli 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with butylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, TETRAETHYLENE GLYCOL | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
4WUT
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE | | Descriptor: | ABC transporter substrate binding protein (Ribose), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE
To be published
|
|
1FIK
 
 | |
1FIL
 
 | |
6DXP
 
 | | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Arcinas, A.J, Ghosh, A, Chamala, S, Bonanno, J.B, Kelly, L, Almo, S.C. | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae
To Be Published
|
|
7JPJ
 
 | | Crystal Structure of the essential dimeric LYSA from Phaeodactylum tricornutum | | Descriptor: | D-LYSINE, Diaminopimelate decarboxylase, SULFATE ION | | Authors: | Fedorov, E, Belinski, V.A, Brunson, J.K, Almo, S.C, Dupont, C.L, Ghosh, A. | | Deposit date: | 2020-08-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Phaeodactylum tricornutum diaminopimelate decarboxylase was acquired via horizontal gene transfer from bacteria and displays substrate promiscuity
Biorxiv, 2020
|
|
6W4L
 
 | | The crystal structure of a single chain H2B-H2A histone chimera from Xenopus laevis | | Descriptor: | Histone H2B 1.1,Histone H2A type 1, PYROPHOSPHATE | | Authors: | Warren, C, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure of a single-chain H2A/H2B dimer.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6WGM
 
 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for pyroglutamate (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, scf7180008839099) in complex with co-purified pyroglutamate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Fedorov, E, Vetting, M.W, Hogle, S.L, Dupont, C.L, Almo, S.C, Ghosh, A. | | Deposit date: | 2020-04-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, GOS_140), in complex with co-purified pyroglutamate
To Be Published
|
|
1ACF
 
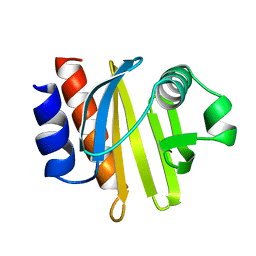 | | ACANTHAMOEBA CASTELLANII PROFILIN IB | | Descriptor: | PROFILIN I | | Authors: | Fedorov, A.A, Magnus, K.A, Graupe, M.H, Lattman, E.E, Pollard, T.D, Almo, S.C. | | Deposit date: | 1994-07-29 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of isoforms of the actin-binding protein profilin that differ in their affinity for phosphatidylinositol phosphates.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1A9T
 
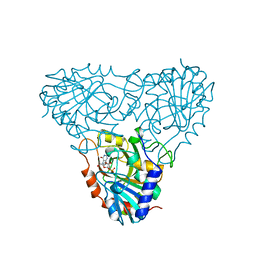 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH 9-DEAZAINOSINE AND PHOSPHATE | | Descriptor: | 1-O-phosphono-alpha-D-ribofuranose, HYPOXANTHINE, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1A9O
 
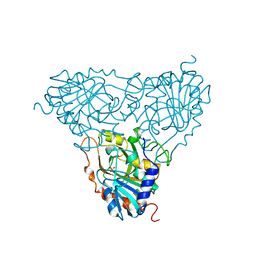 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH PHOSPHATE | | Descriptor: | PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1AAX
 
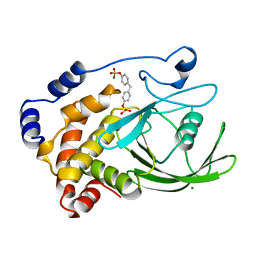 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH TWO BIS(PARA-PHOSPHOPHENYL)METHANE (BPPM) MOLECULES | | Descriptor: | 4-PHOSPHONOOXY-PHENYL-METHYL-[4-PHOSPHONOOXY]BENZEN, MAGNESIUM ION, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Puius, Y.A, Zhao, Y, Sullivan, M, Lawrence, D, Almo, S.C, Zhang, Z.-Y. | | Deposit date: | 1997-01-16 | | Release date: | 1998-03-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1A9P
 
 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH 9-DEAZAINOSINE AND PHOSPHATE | | Descriptor: | 9-DEAZAINOSINE, PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1A9S
 
 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH INOSINE AND SULFATE | | Descriptor: | INOSINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1A9R
 
 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH HYPOXANTHINE AND SULFATE | | Descriptor: | HYPOXANTHINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1A9Q
 
 | | BOVINE PURINE NUCLEOSIDE PHOSPHORYLASE COMPLEXED WITH INOSINE | | Descriptor: | HYPOXANTHINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Mao, C, Cook, W.J, Zhou, M, Fedorov, A.A, Almo, S.C, Ealick, S.E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
6OI0
 
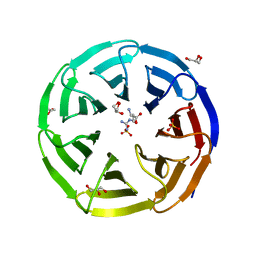 | | Crystal structure of human WDR5 in complex with L-arginine | | Descriptor: | ARGININE, GLYCEROL, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OI3
 
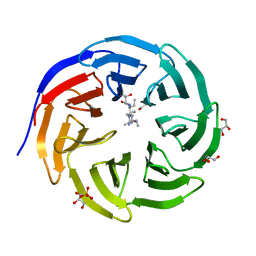 | | Crystal structure of human WDR5 in complex with monomethyl H3R2 peptide | | Descriptor: | GLYCEROL, Monomethyl H3R2 peptide, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OI1
 
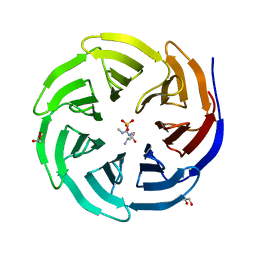 | | Crystal structure of human WDR5 in complex with monomethyl L-arginine | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OI2
 
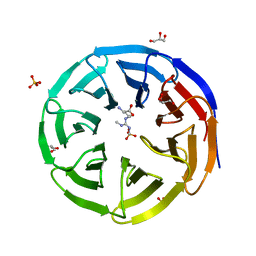 | | Crystal structure of human WDR5 in complex with symmetric dimethyl-L-arginine | | Descriptor: | GLYCEROL, N3, N4-DIMETHYLARGININE, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OFZ
 
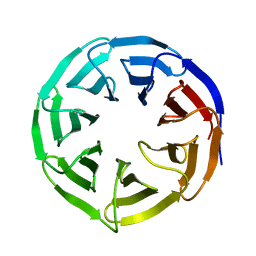 | | Crystal structure of human WDR5 | | Descriptor: | WD repeat-containing protein 5 | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
1CF0
 
 | |
1EEN
 
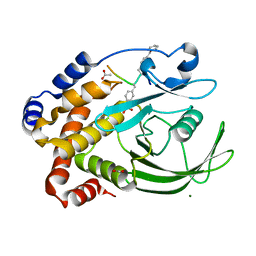 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH ACETYL-D-A-D-BPA-PTYR-L-I-P-Q-Q-G | | Descriptor: | ACETIC ACID, ALA-ASP-PBF-PTR-LEU-ILE-PRO, MAGNESIUM ION, ... | | Authors: | Puius, Y.A, Zhao, Y, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition.
Biochemistry, 39, 2000
|
|
1EEO
 
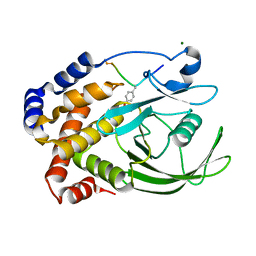 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH ACETYL-E-L-E-F-PTYR-M-D-Y-E-NH2 | | Descriptor: | ACETYL-E-L-E-F-PTYR-M-D-Y-E-NH2 PEPTIDE, MAGNESIUM ION, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Sarmiento, M, Puius, Y.A, Vetter, S.W, Lawrence, D.S, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition.
Biochemistry, 39, 2000
|
|
