5DT2
 
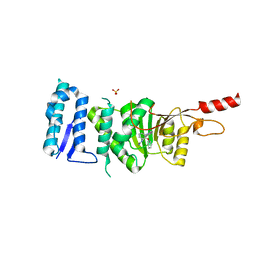 | | Crystal structure of Dot1L in complex with inhibitor CPD11 [N4-methyl-N2-(2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl)pyrimidine-2,4-diamine] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~4~-methyl-N~2~-[2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl]pyrimidine-2,4-diamine, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-17 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
5DRY
 
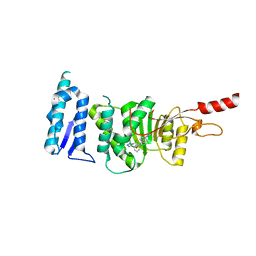 | | Crystal structure of Dot1L in complex with inhibitor CPD3 [N-(1-(2-chlorophenyl)-1H-indol-6-yl)-2-(2-(5-(2-chlorophenyl)-1H-tetrazol-1-yl)acetyl)hydrazinecarboxamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N-[1-(2-chlorophenyl)-1H-indol-6-yl]-2-{[5-(2-chlorophenyl)-1H-tetrazol-1-yl]acetyl}hydrazinecarboxamide, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-16 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
6LGN
 
 | | The atomic structure of varicella zoster virus C-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Li, S, Zheng, Q. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
5B25
 
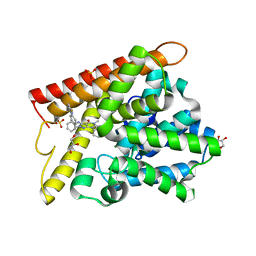 | | Crystal structure of human PDE1B with inhibitor 3 | | Descriptor: | (11R,15S)-4-{[4-(6-fluoropyridin-2-yl)phenyl]methyl}-8-methyl-5-(phenylamino)-1,3,4,8,10-pentaazatetracyclo[7.6.0.02,6.011,15]pentadeca-2,5,9-trien-7-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, GLYCEROL, ... | | Authors: | Ida, K, Lane, W, Snell, G, Sogabe, S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Phosphodiesterase 1 for the Treatment of Cognitive Impairment Associated with Neurodegenerative and Neuropsychiatric Diseases
J.Med.Chem., 59, 2016
|
|
6HQV
 
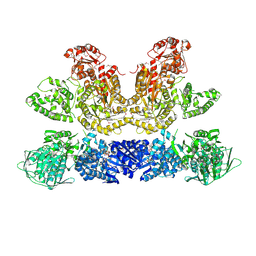 | | Pentafunctional AROM Complex from Chaetomium thermophilum | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, GLUTAMIC ACID, ... | | Authors: | Arora Verasto, H, Hartmann, M.D. | | Deposit date: | 2018-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Architecture and functional dynamics of the pentafunctional AROM complex.
Nat.Chem.Biol., 16, 2020
|
|
7NY0
 
 | |
6Q0D
 
 | | CRYSTAL STRUCTURE OF LDHA IN COMPLEX WITH COMPOUND NCGC00384414-01 AT 2.05 A RESOLUTION | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-[3-(cyclopentylethynyl)-4-fluorophenyl]-5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, GLYCEROL, ... | | Authors: | Dranow, D.M, Davies, D.R. | | Deposit date: | 2019-08-01 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pyrazole-Based Lactate Dehydrogenase Inhibitors with Optimized Cell Activity and Pharmacokinetic Properties.
J.Med.Chem., 63, 2020
|
|
2ZI8
 
 | | Crystal structure of the HsaC extradiol dioxygenase from M. tuberculosis in complex with 3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (DHSA) | | Descriptor: | 3,4-dihydroxy-9,10-secoandrosta-1(10),2,4-triene-9,17-dione, FE (II) ION, PROBABLE BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE BPHC | | Authors: | D'Angelo, I, Yam, K.C, Eltis, L.D, Strynadka, N. | | Deposit date: | 2008-02-13 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis.
Plos Pathog., 5, 2009
|
|
6Q13
 
 | | CRYSTAL STRUCTURE OF LDHA IN COMPLEX WITH COMPOUND NCGC00420737-09 AT 2.00 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-[5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-3-{3-[(5-methylthiophen-2-yl)ethynyl]phenyl}-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, ... | | Authors: | Davies, D.R, Dranow, D.M. | | Deposit date: | 2019-08-02 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyrazole-Based Lactate Dehydrogenase Inhibitors with Optimized Cell Activity and Pharmacokinetic Properties.
J.Med.Chem., 63, 2020
|
|
6PXD
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor, Apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Gouaux, E. | | Deposit date: | 2019-07-25 | | Release date: | 2021-02-10 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
2ZYQ
 
 | | Crystal structure of the HsaC extradiol dioxygenase from M. tuberculosis | | Descriptor: | D(-)-TARTARIC ACID, FE (II) ION, PROBABLE BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE BPHC | | Authors: | D'Angelo, I, Yam, K.C, Eltis, L.D, Strynadka, N. | | Deposit date: | 2009-01-28 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis.
Plos Pathog., 5, 2009
|
|
8EGI
 
 | | X-ray structure of carbonmonoxy hemoglobin in complex with VZHE039-NO | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Donkor, A.K, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-09-12 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Investigation of Novel Nitric Oxide (NO)-Releasing Aromatic Aldehydes as Drug Candidates for the Treatment of Sickle Cell Disease.
Molecules, 27, 2022
|
|
8EII
 
 | | Cryo-EM structure of human DNMT3B homo-tetramer (form II) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EIK
 
 | | Cryo-EM structure of human DNMT3B homo-hexamer | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EIH
 
 | | Cryo-EM structure of human DNMT3B homo-tetramer (form I) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EIJ
 
 | | Cryo-EM structure of human DNMT3B homo-trimer | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
3P43
 
 | | Structure and Activities of Archaeal Members of the LigD 3' Phosphoesterase DNA Repair Enzyme Superfamily | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Smith, P, Nair, P.A, Das, U, Shuman, S. | | Deposit date: | 2010-10-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and activities of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily.
Nucleic Acids Res., 39, 2011
|
|
7CHH
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with BD-368-2 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-368-2 Fab heavy chain, ... | | Authors: | Xiao, J, Zhu, Q, Wang, G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
7CHF
 
 | |
7CHE
 
 | |
7CHB
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with BD-236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-236 Fab heavy chain, BD-236 Fab light chain, ... | | Authors: | Xiao, J, Zhu, Q. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
6IKM
 
 | | Crystal structure of SpuE-Spermidine in complex with ScFv5 | | Descriptor: | Polyamine transport protein, SPERMIDINE, SULFATE ION, ... | | Authors: | Wu, D, Sun, X. | | Deposit date: | 2018-10-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | A Potent Anti-SpuE Antibody Allosterically Inhibits Type III Secretion System and Attenuates Virulence of Pseudomonas Aeruginosa.
J.Mol.Biol., 431, 2019
|
|
4FPW
 
 | | Crystal Structure of CalU16 from Micromonospora echinospora. Northeast Structural Genomics Consortium Target MiR12. | | Descriptor: | CalU16 | | Authors: | Seetharaman, J, Lew, S, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Phillips Jr, G.N, Kennedy, M.A, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-22 | | Release date: | 2012-12-12 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
7YFK
 
 | | The structure of human pregnane X receptor in complex with an SRC-1 coactivator peptide and a limonoid compound, nomilin | | Descriptor: | Nomilin, Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1 | | Authors: | Xia, Y, Yao, D, Huang, C, Cao, Y. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pregnane X receptor agonist nomilin extends lifespan and healthspan in preclinical models through detoxification functions.
Nat Commun, 14, 2023
|
|
