6W5M
 
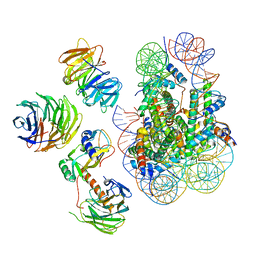 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class02) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5N
 
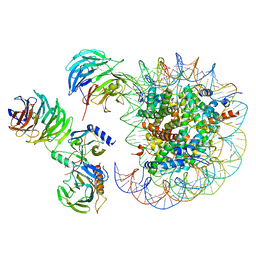 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class05) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6WQ0
 
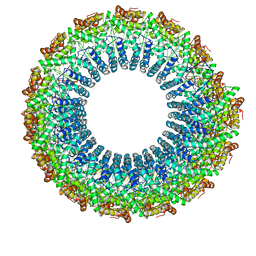 | | Cryo-EM of the S. solfataricus rod-shaped virus, SSRV1 | | Descriptor: | DNA (301-MER), Structural protein | | Authors: | Wang, F, Baquero, D.P, Beltran, L.C, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-29 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6M63
 
 | | Crystal structure of a cAMP sensor G-Flamp1. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Chimera of Cyclic nucleotide-gated potassium channel mll3241 and Yellow fluorescent protein | | Authors: | Zhou, Z, Chen, S, Wang, L, Chu, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging.
Nat Commun, 13, 2022
|
|
8CWM
 
 | |
8CVI
 
 | |
6GMS
 
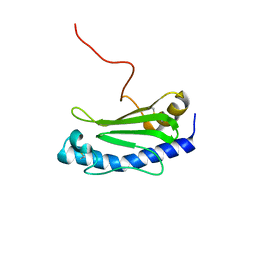 | | Solution NMR structure of the major type IV pilin PpdD from enterohemorrhagic Escherichia coli (EHEC) | | Descriptor: | Prepilin peptidase-dependent protein D | | Authors: | Amorim, G.C, Bardiaux, B, Luna-Rico, A, Zeng, W, Guilvout, I, Egelman, E, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2018-05-28 | | Release date: | 2019-05-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Assembly of the Enterohemorrhagic Escherichia coli Type 4 Pilus.
Structure, 27, 2019
|
|
8CYE
 
 | |
8CXM
 
 | | Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform | | Descriptor: | Flagellin | | Authors: | Sonani, R.R, Kreutzberger, M.A.B, Sebastian, A.L, Scharf, B, Egelman, E.H. | | Deposit date: | 2022-05-21 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergent evolution in the supercoiling of prokaryotic flagellar filaments.
Cell, 185, 2022
|
|
5WZX
 
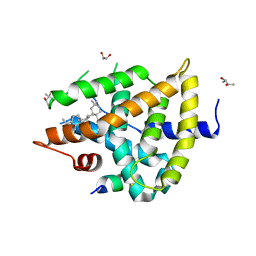 | | Structural basis for a pentacyclic oleanane-type triterpenoid as a ligand of FXR | | Descriptor: | (4aR,6aR,6aS,6bS,8aS,9R,12aR,14bR)-2,2,6a,6b,9,12a-hexamethyl-10-oxidanylidene-1,3,4,5,6,6a,7,8,8a,9,11,12,13,14b-tetradecahydropicene-4a-carboxylic acid, (R,R)-2,3-BUTANEDIOL, Bile acid receptor, ... | | Authors: | Lu, Y, Li, Y. | | Deposit date: | 2017-01-19 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Identification of an Oleanane-Type Triterpene Hedragonic Acid as a Novel Farnesoid X Receptor Ligand with Liver Protective Effects and Anti-inflammatory Activity
Mol. Pharmacol., 93, 2018
|
|
5HZM
 
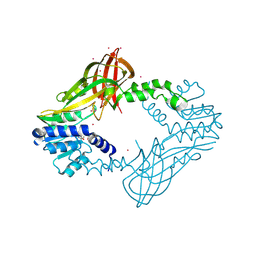 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN ATOM OR ION | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, MIN, J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-02 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
3O4X
 
 | |
3OKQ
 
 | |
3ONX
 
 | |
7CBZ
 
 | | Crystal structure of T2R-TTL-A31 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[5-[4-[2-[4-(2-cyclopropylethanoyl)piperazin-1-yl]ethoxy]phenyl]pyridin-2-yl]-N-(phenylmethyl)ethanamide, CALCIUM ION, ... | | Authors: | Yang, J.H, Yan, W. | | Deposit date: | 2020-06-15 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Design, Synthesis, and Bioactivity Evaluation of Dual-Target Inhibitors of Tubulin and Src Kinase Guided by Crystal Structure.
J.Med.Chem., 64, 2021
|
|
4FIA
 
 | | Crystal Structure of Human CYP46A1 P450 with bicalutamide Bound | | Descriptor: | Cholesterol 24-hydroxylase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Stout, C.D, Pikuleva, I.A, Mast, N. | | Deposit date: | 2012-06-08 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of a cyano- and fluoro-containing drug bicalutamide to cytochrome P450 46A1: unusual features and spectral response.
J.Biol.Chem., 288, 2013
|
|
6IRR
 
 | | Solution structure of DISC1/ATF4 complex | | Descriptor: | Disrupted in schizophrenia 1 homolog,Cyclic AMP-dependent transcription factor ATF-4 | | Authors: | Ye, F, Yu, C, Zhang, M. | | Deposit date: | 2018-11-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural interaction between DISC1 and ATF4 underlying transcriptional and synaptic dysregulation in an iPSC model of mental disorders.
Mol. Psychiatry, 2019
|
|
6JCJ
 
 | | Structure of crolibulin in complex with tubulin | | Descriptor: | (4R)-2,7,8-triamino-4-(3-bromo-4,5-dimethoxyphenyl)-4H-1-benzopyran-3-carbonitrile, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z, Yang, J. | | Deposit date: | 2019-01-29 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of crolibulin in complex with tubulin provides a rationale for drug design.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
4HC4
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
4I51
 
 | | Methyltransferase domain of HUMAN EUCHROMATIC HISTONE METHYLTRANSFERASE 1, mutant Y1211A | | Descriptor: | GLYCEROL, H3K9 NE-ALLYL PEPTIDE, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Dong, A, Zeng, H, Walker, J.R, Islam, K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Lou, M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-11-28 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Defining efficient enzyme-cofactor pairs for bioorthogonal profiling of protein methylation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4OLC
 
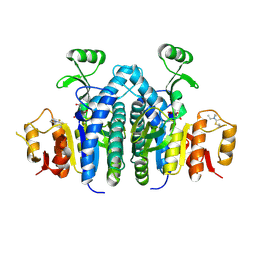 | |
4QQK
 
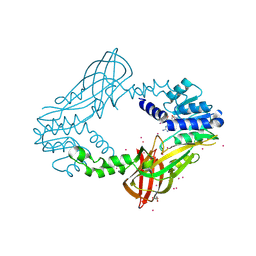 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) with GMS | | Descriptor: | (5S)-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}-N~6~-carbamimidoyl-L-lysine, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
6KNW
 
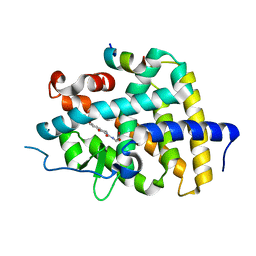 | | THRb mutation with a novel agonist | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
6KNV
 
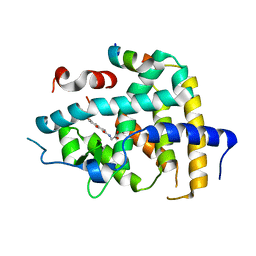 | | THRb mutation with a novel agonist | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
6KKE
 
 | | THRb mutation with a novel agonist | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, SRC2-3, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-07-25 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.577 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
