8SAI
 
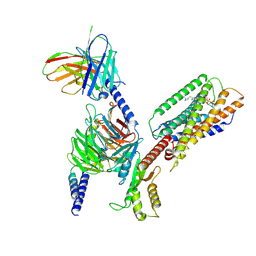 | | Cryo-EM structure of GPR34-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yong, X.H, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2023-04-01 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ZC3
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike
To be published
|
|
8ZC0
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike
To be published
|
|
8ZC1
 
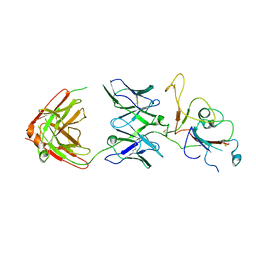 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | Descriptor: | Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, Spike protein S1 | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike
To be published
|
|
8ZC4
 
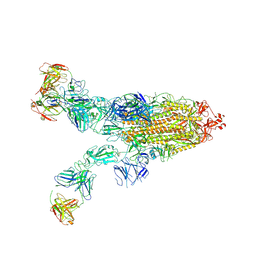 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike
To be published
|
|
8ZBY
 
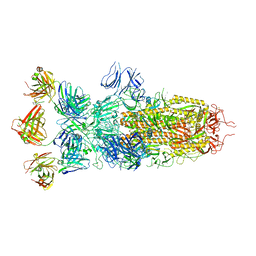 | | SARS-CoV-2 Omicron BA.1 spike trimer (x2-4P) in complex with 3 D1F6 Fabs (0 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike
To be published
|
|
8ZBZ
 
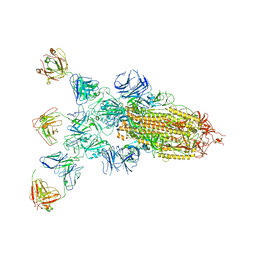 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.71 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike
To be published
|
|
8ZC5
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike
To be published
|
|
8H7L
 
 | | Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7535 fab heavt chain, ... | | Authors: | Liu, Z, Yan, A, Gao, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants.
Nat Commun, 15, 2024
|
|
8H7Z
 
 | |
6PIB
 
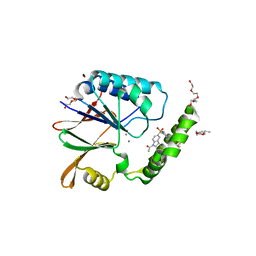 | | Structure of the Klebsiella pneumoniae LpxH-AZ1 complex | | Descriptor: | 1-[5-({4-[3-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-26 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PH9
 
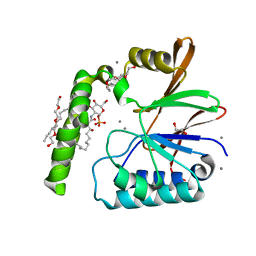 | | Crystal Structure of the Klebsiella pneumoniae LpxH-lipid X complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PJ3
 
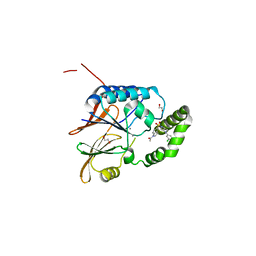 | | Crystal structure of the Klebsiella pneumoniae LpxH/JH-LPH-33 complex | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-({4-[3-chloro-5-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8W9W
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide/phosphoethanolamine | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1, ~{N}-[(~{Z},2~{S},3~{R})-1,3-bis(oxidanyl)heptadec-4-en-2-yl]dodecanamide | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 2024
|
|
8W9Y
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein | | Descriptor: | Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 2024
|
|
7KDT
 
 | |
8WW2
 
 | | GPR3/Gs complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Xiong, Y. | | Deposit date: | 2023-10-24 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Identification of oleic acid as an endogenous ligand of GPR3.
Cell Res., 34, 2024
|
|
7YQE
 
 | |
7TAE
 
 | |
7TAD
 
 | | CryoEM structure of the (NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAC
 
 | | Cryo-EM structure of the (TGA3)2-(NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
6J9D
 
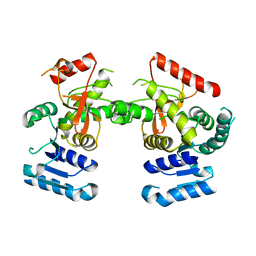 | | Babesia microti lactate dehydrogenase R99A (BmLDHR99A) | | Descriptor: | L-lactate dehydrogenase | | Authors: | Yu, L. | | Deposit date: | 2019-01-22 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Crystal structures ofBabesia microtilactate dehydrogenase BmLDH reveal a critical role for Arg99 in catalysis.
Faseb J., 33, 2019
|
|
6K13
 
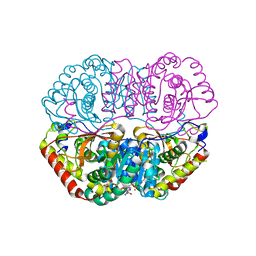 | | Crystal Structure Basis for BmLDH Complex | | Descriptor: | L-lactate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID | | Authors: | Long, Y, Shen, Z. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures ofBabesia microtilactate dehydrogenase BmLDH reveal a critical role for Arg99 in catalysis.
Faseb J., 33, 2019
|
|
6K12
 
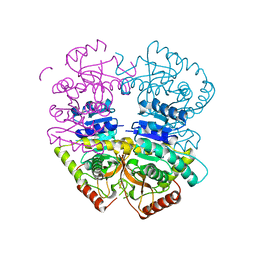 | | Babesia microti lactate dehydrogenase apo form (BmLDH) | | Descriptor: | L-lactate dehydrogenase | | Authors: | Long, Y. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal structures ofBabesia microtilactate dehydrogenase BmLDH reveal a critical role for Arg99 in catalysis.
Faseb J., 33, 2019
|
|
7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
