8HIX
 
 | | Cryo-EM structure of GPR21_m5_Gs | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Chen, B, Lin, X, Xu, F. | | 登録日 | 2022-11-22 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-03-22 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
8HJ2
 
 | | GPR21 wt with G15 complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Chen, B, Lin, X, Xu, F. | | 登録日 | 2022-11-22 | | 公開日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
8HS2
 
 | | Orphan GPR20 in complex with Fab046 | | 分子名称: | Light chain of Fab046, Soluble cytochrome b562,G-protein coupled receptor 20, heavy chain of Fab046 | | 著者 | Lin, X, Jiang, S, Xu, F. | | 登録日 | 2022-12-16 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HJ1
 
 | | GPR21(wt) and Gs complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Chen, B, Lin, X, Xu, F. | | 登録日 | 2022-11-22 | | 公開日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
8HQ2
 
 | |
8HQ1
 
 | |
8HPW
 
 | |
8HPY
 
 | | Crystal structure of human LGI1-ADAM22 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Disintegrin and metalloproteinase domain-containing protein 22, ... | | 著者 | Liu, H, Xu, F. | | 登録日 | 2022-12-13 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (5.87 Å) | | 主引用文献 | Crystal structure of human LGI1-ADAM22 complex
To Be Published
|
|
8HPX
 
 | |
5HKE
 
 | | bile salt hydrolase from Lactobacillus salivarius | | 分子名称: | Bile salt hydrolase, PHOSPHATE ION | | 著者 | Hu, X.-J. | | 登録日 | 2016-01-14 | | 公開日 | 2016-05-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of bile salt hydrolase from Lactobacillus salivarius.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5Y7P
 
 | |
1O5T
 
 | | Crystal structure of the aminoacylation catalytic fragment of human tryptophanyl-tRNA synthetase | | 分子名称: | Tryptophanyl-tRNA synthetase | | 著者 | Yu, Y, Liu, Y, Shen, N, Xu, X, Jia, J, Jin, Y, Arnold, E, Ding, J. | | 登録日 | 2003-10-05 | | 公開日 | 2004-07-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of Human Tryptophanyl-tRNA Synthetase Catalytic Fragment
J.BIOL.CHEM., 279, 2004
|
|
8W8Q
 
 | | Cryo-EM structure of the GPR101-Gs complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Sun, J.P, Gao, N, Yu, X, Wang, G.P, Yang, F, Wang, J.Y, Yang, Z, Guan, Y. | | 登録日 | 2023-09-04 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8S
 
 | | Cryo-EM structure of the AA14-bound GPR101 complex | | 分子名称: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Probable G-protein coupled receptor 101 | | 著者 | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | 登録日 | 2023-09-04 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8R
 
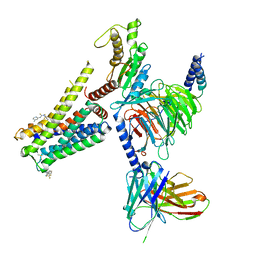 | | Cryo-EM structure of the AA-14-bound GPR101-Gs complex | | 分子名称: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | 登録日 | 2023-09-04 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
7LKC
 
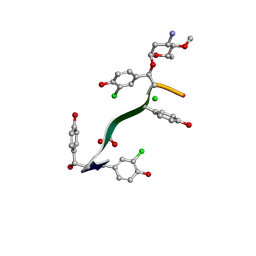 | | Crystal Structure of Keratinimicin A | | 分子名称: | (2~{S},4~{S},5~{R},6~{S})-4-azanyl-5-methoxy-6-methyl-oxan-2-ol, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Davis, K.M, Jeffrey, P.D, Seyedsayamdost, M.R. | | 登録日 | 2021-02-02 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Structural and Functional Analysis of Keratinicyclin Reveals Synergistic Antibiosis with Vancomycin against Clostridium difficile
to be published
|
|
7LTB
 
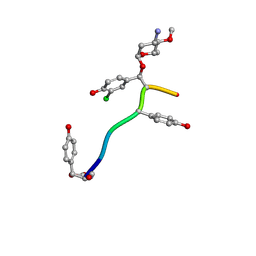 | | Crystal Structure of Keratinicyclin B | | 分子名称: | (2~{S},4~{S},5~{R},6~{S})-4-azanyl-5-methoxy-6-methyl-oxan-2-ol, 3-ammonio-2,3,6-trideoxy-alpha-L-arabino-hexopyranose-(1-2)-beta-D-glucopyranose, FORMIC ACID, ... | | 著者 | Davis, K.M, Jeffrey, P.D, Seyedsayamdost, M.R. | | 登録日 | 2021-02-19 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Structural and Functional Analysis of Keratinicyclin Reveals Synergistic Antibiosis with Vancomycin against Clostridium difficile
to be published
|
|
8E9E
 
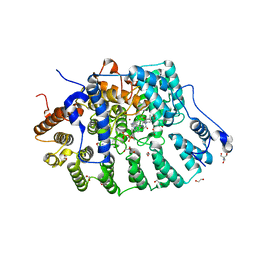 | | Rat protein farnesyltransferase in complex with FPP and inhibitor 2f | | 分子名称: | (5S)-4-({1-[(4-bromophenyl)methyl]-1H-imidazol-5-yl}methyl)-5-butyl-1-[3-(trifluoromethoxy)phenyl]piperazin-2-one, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Wang, Y, Shi, Y, Beese, L.S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.844 Å) | | 主引用文献 | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
3J1P
 
 | |
6KCW
 
 | | Structure of alginate lyase Aly36B | | 分子名称: | Alginate lyase, CALCIUM ION, PHOSPHATE ION | | 著者 | Dong, F, Chen, X.L, Zhang, Y.Z. | | 登録日 | 2019-06-29 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Alginate Lyase Aly36B is a New Bacterial Member of the Polysaccharide Lyase Family 36 and Catalyzes by a Novel Mechanism With Lysine as Both the Catalytic Base and Catalytic Acid.
J.Mol.Biol., 431, 2019
|
|
6KHJ
 
 | | Supercomplex for electron transfer | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | 登録日 | 2019-07-15 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
6SJ7
 
 | |
6KHI
 
 | | Supercomplex for cylic electron transport in cyanobacteria | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | 登録日 | 2019-07-15 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
6KCV
 
 | |
6KZK
 
 | |
