6V3F
 
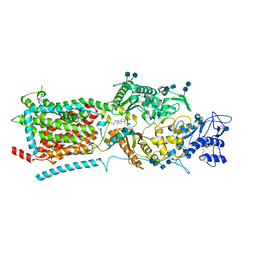 | | Structure of NPC1-like intracellular cholesterol transporter 1 (NPC1L1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Huang, C.S, Yu, X, Min, X, Wang, Z. | | Deposit date: | 2019-11-25 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of NPC1L1 reveal mechanisms of cholesterol transport and ezetimibe inhibition
Sci Adv, 6, 2020
|
|
6V3H
 
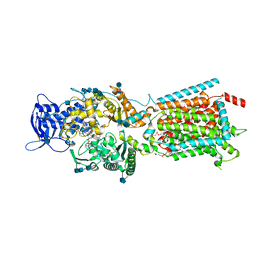 | | Structure of NPC1-like intracellular cholesterol transporter 1 (NPC1L1) in complex with an ezetimibe analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(2S,3R)-3-[(3S)-3-(4-fluorophenyl)-3-hydroxypropyl]-1-(4-{3-[(methylsulfonyl)amino]prop-1-yn-1-yl}phenyl)-4-oxoazetidin-2-yl]phenyl beta-D-glucopyranosiduronic acid, ... | | Authors: | Huang, C.S, Yu, X, Min, X, Wang, Z. | | Deposit date: | 2019-11-25 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of NPC1L1 reveal mechanisms of cholesterol transport and ezetimibe inhibition
Sci Adv, 6, 2020
|
|
5UD8
 
 | | Crystal Structure of Mutant Ig-like Domain | | Descriptor: | Triggering receptor expressed on myeloid cells 2 | | Authors: | Sudom, A, Min, X, Wang, Z. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-04 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
5UD7
 
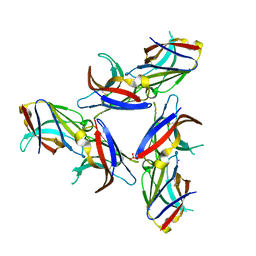 | | Crystal Structure of Wild-Type Ig-like Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IODIDE ION, SULFATE ION, ... | | Authors: | Sudom, A, Min, X, Wang, Z. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.20002246 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
7JR7
 
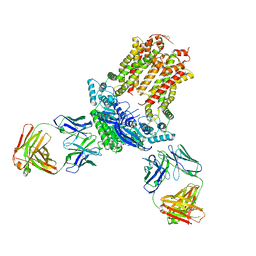 | | Cryo-EM structure of ABCG5/G8 in complex with Fab 2E10 and 11F4 | | Descriptor: | ATP-binding cassette sub-family G member 5, ATP-binding cassette sub-family G member 8, Fab 11F4 heavy chain, ... | | Authors: | Huang, C.S, Yu, X, Min, X, Wang, Z, Zhang, H. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-07 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of ABCG5/G8 in complex with modulating antibodies
Commun Biol, 4, 2021
|
|
4FFV
 
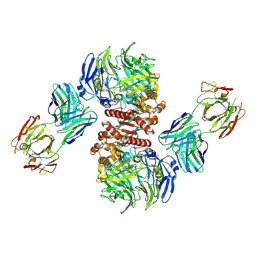 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with 11A19 Fab | | Descriptor: | 11A19 Fab heavy chain, 11A19 Fab light chain, Dipeptidyl peptidase 4 | | Authors: | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | Deposit date: | 2012-06-01 | | Release date: | 2012-12-12 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
3P4K
 
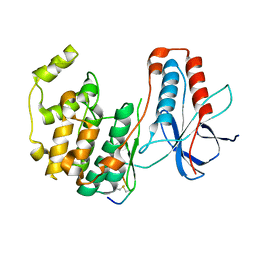 | | The third conformation of p38a MAP kinase observed in phosphorylated p38a and in solution | | Descriptor: | MAP kinase 14, Mitogen-activated protein kinase 14 | | Authors: | Akella, R, Min, X, Wu, Q, Gardner, K.H, Goldsmith, E.J. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | The third conformation of p38a MAP kinase observed in phosphorylated p38a and in solution
Structure, 18, 2010
|
|
6CN9
 
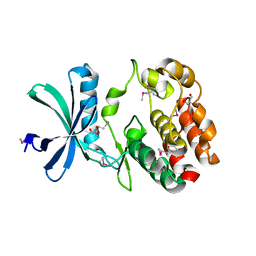 | | Crystal structure of the Kinase domain of WNK1 | | Descriptor: | GLYCEROL, SULFATE ION, Serine/threonine-protein kinase WNK1 | | Authors: | Akella, R, Goldsmith, E.J. | | Deposit date: | 2018-03-07 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension.
Structure, 12, 2004
|
|
8TJC
 
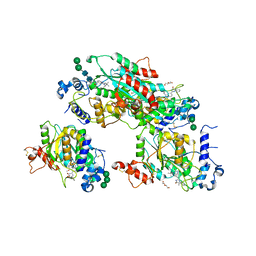 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 8a | | Descriptor: | (6M)-1-[(2R)-3,3-dimethylbutan-2-yl]-6-[(5S)-5-methyl-4-oxo-5-phenyl-4,5-dihydro-1H-imidazol-2-yl]-1,3-dihydro-2H-benzimidazol-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-07-20 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
5IVT
 
 | | Crystal Structure of HIV Protease complexed with [(1S)-1-[(S)-(4-chlorophenyl)-(3,5-difluorophenyl)methyl]-2-[[5-fluoro-4-[2-[(2R,5S)-5-(2,2,2-trifluoroethylcarbamoyloxymethyl)morpholin-4-ium-2-yl]ethyl]pyridin-1-ium-3-yl]amino]-2-oxo-ethyl]ammonium | | Descriptor: | (betaS)-4-chloro-beta-(3,5-difluorophenyl)-N-(5-fluoro-4-{2-[(2R,5S)-5-({[(2,2,2-trifluoroethyl)carbamoyl]oxy}methyl)morpholin-2-yl]ethyl}pyridin-3-yl)-L-phenylalaninamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Su, H.P. | | Deposit date: | 2016-03-21 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery of MK-8718, an HIV Protease Inhibitor Containing a Novel Morpholine Aspartate Binding Group.
Acs Med.Chem.Lett., 7, 2016
|
|
5IVR
 
 | |
5IVS
 
 | |
8SZ3
 
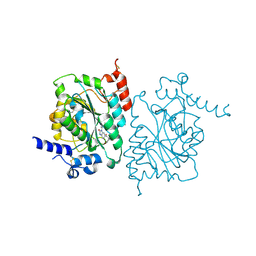 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 7j | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(1S)-1-(5-bromopyridin-2-yl)ethyl]-3-[(2R)-3,3-dimethylbutan-2-yl]-2-oxo-2,3-dihydro-1H-benzimidazole-5-carboxamide, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-05-26 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
8TIC
 
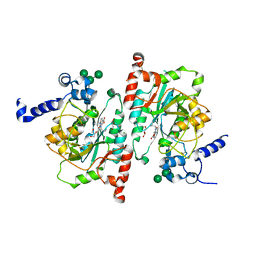 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-methylpropyl)-2-oxo-2,3-dihydro-1H-benzimidazole-5-carboxylic acid, CHLORIDE ION, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-07-19 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
5IVQ
 
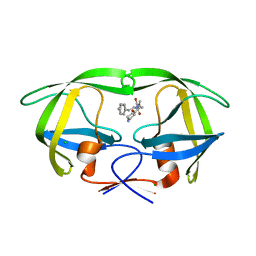 | |
6B8O
 
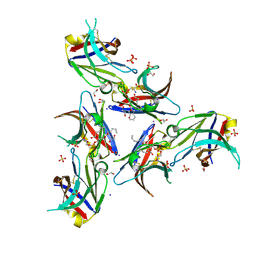 | | WT Ig-like V Domain with Phosphatidylserine | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sudom, A, Wang, Z. | | Deposit date: | 2017-10-09 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
5X56
 
 | | Crystal structure of PSB27 from Arabidopsis thaliana | | Descriptor: | Photosystem II repair protein PSB27-H1, chloroplastic | | Authors: | Liu, J, Cheng, X. | | Deposit date: | 2017-02-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Psb27 from Arabidopsis thaliana determined at a resolution of 1.85 angstrom.
Photosyn. Res., 136, 2018
|
|
5XFJ
 
 | | Crystal structure of LY2874455 in complex of FGFR4 gatekeeper mutation (V550M) | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance.
Chem. Commun. (Camb.), 54, 2018
|
|
5XFF
 
 | | Crystal structure of LY2874455 in complex of FGFR4 gatekeeper mutation (V550L) | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance.
Chem. Commun. (Camb.), 54, 2018
|
|
5VTA
 
 | | Co-Crystal Structure of DPPIV with a Chemibody Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{4-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl]piperazin-1-yl}-N-(22-oxo-3,6,9,12,15,18-hexaoxa-21-azatricosan-1-yl)acetamide, ... | | Authors: | Wang, Z, Johnstone, S, Cheng, A. | | Deposit date: | 2017-05-16 | | Release date: | 2018-05-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-guided Discovery of Dual-recognition Chemibodies.
Sci Rep, 8, 2018
|
|
6B3F
 
 | |
6B38
 
 | |
6B3G
 
 | |
4FF5
 
 | |
6B36
 
 | |
