6JLS
 
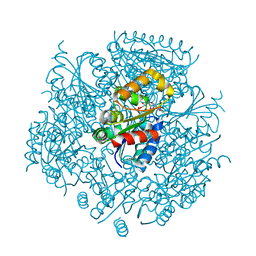 | | Crystal Structure of FMN-dependent Cysteine Decarboxylases TvaF from Thioviridamide Biosynthesis | | Descriptor: | FLAVIN MONONUCLEOTIDE, Putative flavoprotein decarboxylase | | Authors: | Li, J, Lu, J, Wang, H, Zhu, J. | | Deposit date: | 2019-03-06 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of the FMN-Dependent Cysteine Decarboxylase from Thioviridamide Biosynthesis.
Org.Lett., 21, 2019
|
|
8EC7
 
 | | HnRNPA2 D290V LCD PM3 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
7SMV
 
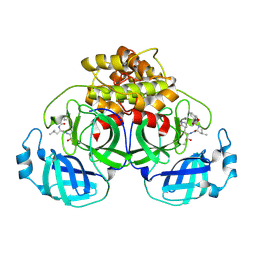 | | Crystallization of feline coronavirus Mpro with GC376 reveals mechanism of inhibition | | Descriptor: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Khan, M.B, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-10-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallization of Feline Coronavirus M pro With GC376 Reveals Mechanism of Inhibition.
Front Chem, 10, 2022
|
|
7SNA
 
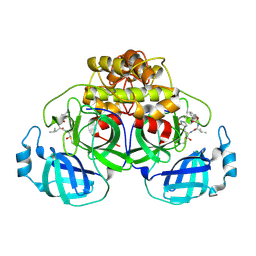 | | Crystallization of feline coronavirus Mpro with GC376 reveals mechanism of inhibition | | Descriptor: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Khan, M.B, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallization of Feline Coronavirus M pro With GC376 Reveals Mechanism of Inhibition.
Front Chem, 10, 2022
|
|
7LUX
 
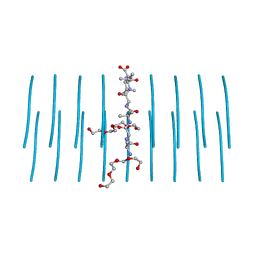 | | AALALL segment from the Nucleoprotein of SARS-CoV-2, residues 217-222, crystal form 2 | | Descriptor: | Nucleoprotein AALALL, TETRAETHYLENE GLYCOL | | Authors: | Lu, J, Zee, C.-T, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.303 Å) | | Cite: | Inhibition of amyloid formation of the Nucleoprotein of SARS-CoV-2.
Biorxiv, 2021
|
|
5V71
 
 | | KRAS G12C in bound to quinazoline based switch II pocket (SWIIP) binder | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Westover, K, Lu, J. | | Deposit date: | 2017-03-17 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.228 Å) | | Cite: | KRAS G12C Drug Development: Discrimination between Switch II Pocket Configurations Using Hydrogen/Deuterium-Exchange Mass Spectrometry.
Structure, 25, 2017
|
|
6ASE
 
 | | KRAS mutant-A59G in GDP-bound | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Westover, K, Lu, J. | | Deposit date: | 2017-08-24 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.554 Å) | | Cite: | KRAS Switch Mutants D33E and A59G Crystallize in the State 1 Conformation.
Biochemistry, 57, 2018
|
|
6BP1
 
 | |
6NOV
 
 | | A Fab derived from ixekizumab | | Descriptor: | DODECAETHYLENE GLYCOL, Fab Heavy Chain, Fab Light Chain | | Authors: | Durbin, J.D, Clawson, D.K, Lu, F, Tian, Y, Lu, J, Schmitt, M, Atwell, S. | | Deposit date: | 2019-01-16 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Development of tibulizumab, a tetravalent bispecific antibody targeting BAFF and IL-17A for the treatment of autoimmune disease.
Mabs, 11, 2019
|
|
8T1U
 
 | | Crystal structure of the DRM2-CTA DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*AP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*TP*TP*TP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Chen, J, Lu, J, Song, J. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | DNA conformational dynamics in the context-dependent non-CG CHH methylation by plant methyltransferase DRM2.
J.Biol.Chem., 299, 2023
|
|
6NOU
 
 | | An scFv derived from ixekizumab | | Descriptor: | GLYCEROL, scFv derived from ixekizumab | | Authors: | Durbin, J.D, Clawson, D.K, Lu, F, Tian, Y, Lu, J, Atwell, S. | | Deposit date: | 2019-01-16 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Development of tibulizumab, a tetravalent bispecific antibody targeting BAFF and IL-17A for the treatment of autoimmune disease.
Mabs, 11, 2019
|
|
5MEI
 
 | | Crystal structure of Agelastatin A bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | McClary, B, Zinshteyn, B, Meyer, M, Jouanneau, M, Pellegrino, S, Yusupova, G, Schuller, A, Reyes, J.C.P, Lu, J, Luo, C, Dang, Y, Romo, D, Yusupov, M, Green, R, Liu, J.O. | | Deposit date: | 2016-11-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of Eukaryotic Translation by the Antitumor Natural Product Agelastatin A.
Cell Chem Biol, 24, 2017
|
|
5KYK
 
 | | Covalent GTP-competitive inhibitors of KRAS G12C: Guanosine bisphosphonate Analogs | | Descriptor: | 5'-O-[(R)-[({2-[(chloroacetyl)amino]ethyl}sulfamoyl)methyl](hydroxy)phosphoryl]guanosine, GTPase KRas | | Authors: | Xiong, Y, Lu, J, Hunter, J, Li, L, Scott, D, Manandhar, A, Gondi, S, Westover, K.D, Gray, N.S. | | Deposit date: | 2016-07-21 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Covalent Guanosine Mimetic Inhibitors of G12C KRAS.
ACS Med Chem Lett, 8, 2017
|
|
1F03
 
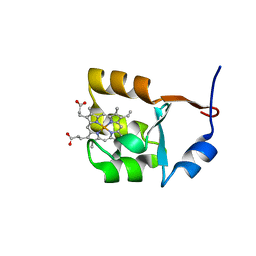 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
8T12
 
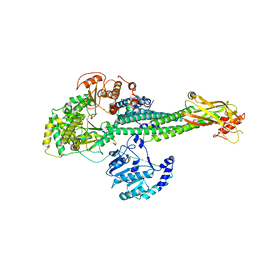 | |
8T13
 
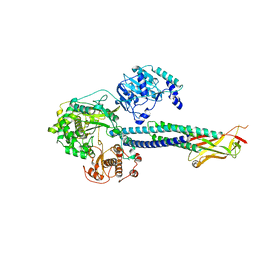 | |
2WZZ
 
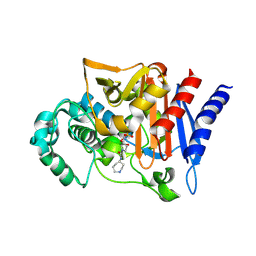 | |
2WZX
 
 | |
6NYT
 
 | | Munc13-1 C2B-domain, calcium bound | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tomchick, D.R, Rizo, J, Machius, M, Lu, J. | | Deposit date: | 2019-02-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis.
Nat. Struct. Mol. Biol., 17, 2010
|
|
6NYC
 
 | | Munc13-1 C2B-domain, calcium free | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Munc13-1 | | Authors: | Tomchick, D.R, Rizo, J, Machius, M, Lu, J. | | Deposit date: | 2019-02-11 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis.
Nat. Struct. Mol. Biol., 17, 2010
|
|
2A20
 
 | | Solution structure of Rim2 Zinc Finger Domain | | Descriptor: | Regulating synaptic membrane exocytosis protein 2, ZINC ION | | Authors: | Dulubova, I, Lou, X, Lu, J, Huryeva, I, Alam, A, Schneggenburger, R, Sudhof, T.C, Rizo, J. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A Munc13/RIM/Rab3 tripartite complex: from priming to plasticity?
Embo J., 24, 2005
|
|
1ZHA
 
 | | A. aeolicus KDO8PS R106G mutant in complex with PEP and R5P | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Xu, X, Kona, F, Wang, J, Lu, J, Stemmler, T, Gatti, D.L. | | Deposit date: | 2005-04-25 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Catalytic and Conformational Cycle of Aquifex aeolicus KDO8P Synthase: Role of the L7 Loop.
Biochemistry, 44, 2005
|
|
1ZJI
 
 | | Aquifex aeolicus KDO8PS R106G mutant in complex with 2PGA and R5P | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, ... | | Authors: | Xu, X, Kona, F, Wang, J, Lu, J, Stemmler, T, Gatti, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Catalytic and Conformational Cycle of Aquifex aeolicus KDO8P Synthase: Role of the L7 Loop
Biochemistry, 44, 2005
|
|
4QPO
 
 | | Mechanistic basis of plasmid-specific DNA binding of the F plasmid regulatory protein, TraM | | Descriptor: | PHOSPHATE ION, Relaxosome protein TraM | | Authors: | Peng, Y, Lu, J, Wong, J, Edwards, R.A, Frost, L.S, Glover, J.N.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Mechanistic Basis of Plasmid-Specific DNA Binding of the F Plasmid Regulatory Protein, TraM.
J.Mol.Biol., 426, 2014
|
|
