7NUH
 
 | |
6G02
 
 | | Complex of neuraminidase from H1N1 influenza virus with tamiphosphor omega-azidohexyl ester | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[[(3~{R},4~{R},5~{S})-4-acetamido-5-azanyl-3-pentan-3-yloxy-cyclohexen-1-yl]-oxidanyl-phosphoryl]oxyhexylimino-azanylidene-azanium, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | DNA-linked inhibitor antibody assay (DIANA) as a new method for screening influenza neuraminidase inhibitors.
Biochem. J., 475, 2018
|
|
7AS0
 
 | | Influenza A PB2 in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, BROMIDE ION, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS1
 
 | | Influenza A PB2 (F404Y mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, CHLORIDE ION, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS2
 
 | | Influenza A PB2 (M431 mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS3
 
 | | Influenza A PB2 (H357N mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
4Z2Z
 
 | |
6RZX
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor FBSA | | Descriptor: | 1,1,2,2,3,3,4,4,4-nonakis(fluoranyl)butane-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
6S03
 
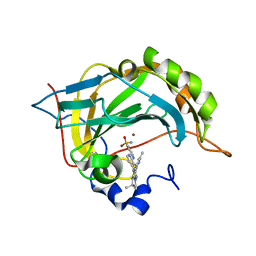 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor I39LT379 | | Descriptor: | 4-[[4-[5,5-dimethyl-2-(6-methylpyridin-2-yl)-4,6-dihydropyrrolo[1,2-b]pyrazol-3-yl]pyridin-2-yl]amino]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
1M0B
 
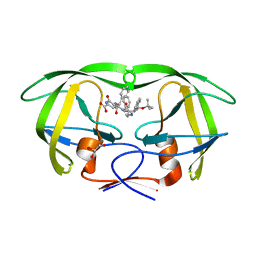 | | HIV-1 protease in complex with an ethyleneamine inhibitor | | Descriptor: | GLYCEROL, N-{(3S)-3-[(tert-butoxycarbonyl)amino]-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Petrokova, H, Hasek, J, Dohnalek, J. | | Deposit date: | 2002-06-12 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Role of hydroxyl group and R/S configuration of isostere in binding properties of HIV-1 protease inhibitors
Eur.J.Biochem., 271, 2004
|
|
1ZSF
 
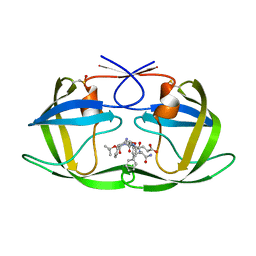 | | Crystal Structure of Complex of a Hydroxyethylamine Inhibitor with HIV-1 Protease at 2.0A Resolution | | Descriptor: | N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Duskova, J, Skalova, T, Dohnalek, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-24 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | On the role of the R configuration of the reaction-intermediate isostere in HIV-1 protease-inhibitor binding: X-ray structure at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1ZSR
 
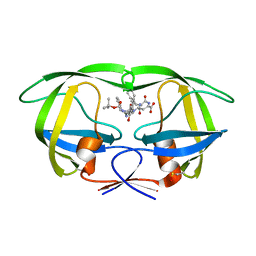 | | Crystal structure of wild type HIV-1 protease (BRU isolate) with a hydroxyethylamine peptidomimetic inhibitor BOC-PHE-PSI[S-CH(OH)CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | N-{(2S,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-24 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | On the role of the R configuration of the reaction-intermediate isostere in HIV-1 protease-inhibitor binding: X-ray structure at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6GXB
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor JS13 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, ~{N}-[4-[4-[(4-sulfamoylphenyl)carbamoylamino]phenoxy]butyl]ethanamide | | Authors: | Brynda, J, Rezacova, P, Pospisilova, K. | | Deposit date: | 2018-06-27 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibitor-Polymer Conjugates as a Versatile Tool for Detection and Visualization of Cancer-Associated Carbonic Anhydrase Isoforms
Acs Omega, 4, 2019
|
|
6GXE
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor JS14 | | Descriptor: | 3-[2-[2-[2-[2-(aminomethyloxy)ethoxy]ethoxy]ethoxy]ethoxy]-~{N}-[4-[4-[(4-sulfamoylphenyl)carbamoylamino]phenoxy]butyl]propanamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Brynda, J, Rezacova, P, Pospisilova, K. | | Deposit date: | 2018-06-27 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Inhibitor-Polymer Conjugates as a Versatile Tool for Detection and Visualization of Cancer-Associated Carbonic Anhydrase Isoforms
Acs Omega, 2019
|
|
6HP0
 
 | | Complex of Neuraminidase from H1N1 Influenza Virus in Complex with Oseltamivir Triazol Derivative | | Descriptor: | (3~{R},4~{R},5~{S})-4-acetamido-5-[4-(hydroxymethyl)-1,2,3-triazol-1-yl]-3-pentan-3-yloxy-cyclohexene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2018-09-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Investigation of flexibility of neuraminidase 150-loop using tamiflu derivatives in influenza A viruses H1N1 and H5N1.
Bioorg.Med.Chem., 27, 2019
|
|
1ZJ7
 
 | | Crystal structure of a complex of mutant HIV-1 protease (A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor BOC-PHE-PSI[S-CH(OH)CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | N-{(2S,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-04-28 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | HIV-1 protease mutations and inhibitor modifications monitored on a series of complexes. Structural basis for the effect of the A71V mutation on the active site
J.Med.Chem., 49, 2006
|
|
1ZLF
 
 | | Crystal structure of a complex of mutant HIV-1 protease (A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor | | Descriptor: | N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-glutaminyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-06 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HIV-1 protease mutations and inhibitor modifications monitored on a series of complexes. Structural basis for the effect of the A71V mutation on the active site
J.Med.Chem., 49, 2006
|
|
3BI0
 
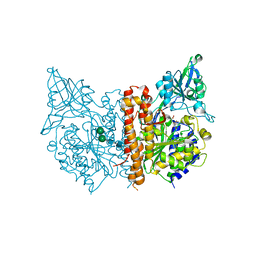 | |
3BI1
 
 | |
3BHX
 
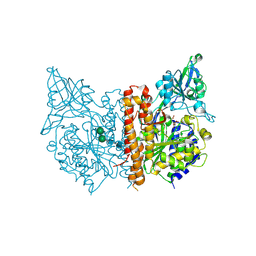 | |
3BXM
 
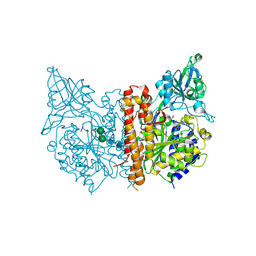 | | Structure of an inactive mutant of human glutamate carboxypeptidase II [GCPII(E424A)] in complex with N-acetyl-Asp-Glu (NAAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lubkowski, J, Barinka, C. | | Deposit date: | 2008-01-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Reaction mechanism of glutamate carboxypeptidase II revealed by mutagenesis, X-ray crystallography, and computational methods.
Biochemistry, 48, 2009
|
|
5KES
 
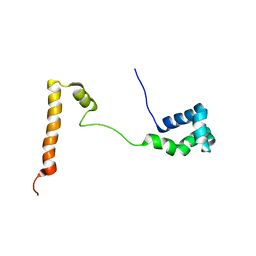 | | Solution structure of the yeast Ddi1 HDD domain | | Descriptor: | DNA damage-inducible protein 1 | | Authors: | Trempe, J.-F, Ratcliffe, C, Veverka, V, Saskova, K, Gehring, K. | | Deposit date: | 2016-06-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the yeast DNA damage-inducible protein Ddi1 reveal domain architecture of this eukaryotic protein family.
Sci Rep, 6, 2016
|
|
3FF3
 
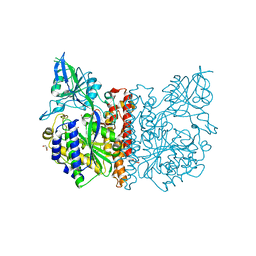 | |
3FEC
 
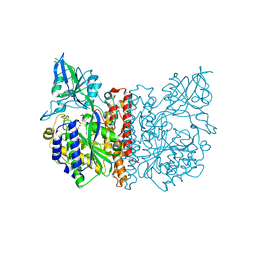 | | Crystal structure of human Glutamate Carboxypeptidase III (GCPIII/NAALADase II), pseudo-unliganded | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Barinka, C, Lubkowski, J, Hlouchova, K. | | Deposit date: | 2008-11-28 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III
Febs J., 276, 2009
|
|
3FEE
 
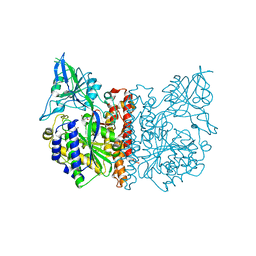 | | The high resolution structure of human glutamate carboxypeptidase III (GCPIII/NAALADase II) in complex with quisqualic acid | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubkowski, J, Barinka, C, Hlouchova, K. | | Deposit date: | 2008-11-28 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III
Febs J., 276, 2009
|
|
