3RSX
 
 | |
3RTM
 
 | |
3RU1
 
 | |
3RVI
 
 | |
7DCN
 
 | | Apo-citrate lyase phosphoribosyl-dephospho-CoA transferase | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, SULFATE ION, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
7DCM
 
 | | Crystal structure of CITX | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
4W8F
 
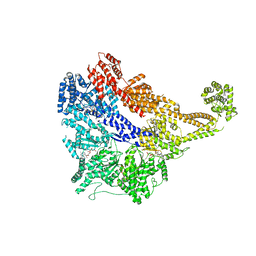 | | Crystal structure of the dynein motor domain in the AMPPNP-bound state | | Descriptor: | Dynein heavy chain lysozyme chimera, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Cheng, H.-C, Bhabha, G, Zhang, N, Vale, R.D. | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.541 Å) | | Cite: | Allosteric communication in the Dynein motor domain.
Cell, 159, 2014
|
|
4DUS
 
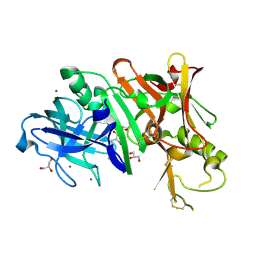 | | Structure of Bace-1 (Beta-Secretase) in complex with N-((2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-((6'-neopentyl-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl)amino)butan-2-yl)acetamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-10 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Potent and Orally Efficacious, Hydroxyethylamine-Based Inhibitor of beta-Secretase.
ACS Med Chem Lett, 3, 2012
|
|
4XKX
 
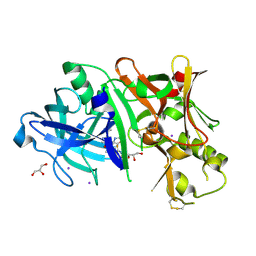 | | Crystal structure of BACE1 in complex with 2-aminooxazoline 3-azaxanthene inhibitor 28 | | Descriptor: | (5S)-7-(2-fluoropyridin-3-yl)-3-(2-fluoropyridin-4-yl)spiro[chromeno[2,3-c]pyridine-5,4'-[1,3]oxazol]-2'-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-01-12 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of 2-aminooxazoline 3-azaxanthenes as orally efficacious beta-secretase inhibitors for the potential treatment of Alzheimer's disease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8CVT
 
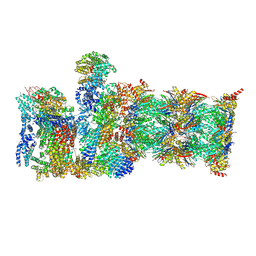 | | Human 19S-20S proteasome, state SD2 | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 8, 26S proteasome complex subunit SEM1, ... | | Authors: | Zhao, J. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8CVR
 
 | | Human 20S proteasome with MG-132 | | Descriptor: | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-4-methyl-1-oxopentan-2-yl]-L-leucinamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Zhao, J. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8CVS
 
 | | Human PA200-20S proteasome with MG-132 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-4-methyl-1-oxopentan-2-yl]-L-leucinamide, Proteasome activator complex subunit 4, ... | | Authors: | Zhao, J. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6SCT
 
 | | Cryo-EM structure of the consensus triskelion hub of the clathrin coat complex | | Descriptor: | Clathrin heavy chain, Clathrin light chain | | Authors: | Morris, K.L, Cameron, A.D, Sessions, R, Smith, C.J. | | Deposit date: | 2019-07-25 | | Release date: | 2019-10-02 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Cryo-EM of multiple cage architectures reveals a universal mode of clathrin self-assembly.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2MLK
 
 | | Three-dimensional structure of the C-terminal DNA-binding domain of RstA protein from Klebsiella pneumoniae | | Descriptor: | RstA | | Authors: | Fang, P, Chen, S, Cheng, Y, Chang, C, Yu, T, Huang, T. | | Deposit date: | 2014-03-02 | | Release date: | 2014-07-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element.
Nucleic Acids Res., 42, 2014
|
|
7T32
 
 | | CryoEM structure of the adenosine 2A receptor-BRIL/Anti BRIL Fab complex with ZM241385 | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 Fusion Protein | | Authors: | Zhang, K.H, Wu, H, Hoppe, N, Manglik, A, Cheng, Y.F. | | Deposit date: | 2021-12-06 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Fusion protein strategies for cryo-EM study of G protein-coupled receptors.
Nat Commun, 13, 2022
|
|
3J9J
 
 | | Structure of the capsaicin receptor, TRPV1, determined by single particle electron cryo-microscopy | | Descriptor: | Transient receptor potential cation channel subfamily V member 1 | | Authors: | Wang, R.Y.-R, Barad, B.A, Fraser, J.S, DiMaio, F. | | Deposit date: | 2015-02-02 | | Release date: | 2015-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.275 Å) | | Cite: | EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy.
Nat.Methods, 12, 2015
|
|
3EFK
 
 | | Structure of c-Met with pyrimidone inhibitor 50 | | Descriptor: | 5-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-2-[(4-fluorophenyl)amino]-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, D'Angelo, N, Whittington, D, Dussault, I. | | Deposit date: | 2008-09-09 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and biological evaluation of potent c-Met inhibitors.
J.Med.Chem., 51, 2008
|
|
3EFJ
 
 | | Structure of c-Met with pyrimidone inhibitor 7 | | Descriptor: | 2-benzyl-5-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | D'Angelo, N, Bellon, S, Whittington, D. | | Deposit date: | 2008-09-09 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, and biological evaluation of potent c-Met inhibitors.
J.Med.Chem., 51, 2008
|
|
7C02
 
 | | Crystal structure of dimeric MERS-CoV receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Dai, L, Qi, J, Gao, G.F. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS.
Cell, 182, 2020
|
|
3BWN
 
 | | L-tryptophan aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-tryptophan aminotransferase, PHENYLALANINE, ... | | Authors: | Ferrer, J.-L, Noel, J.P, Pojer, F, Bowman, M, Chory, J, Tao, Y. | | Deposit date: | 2008-01-10 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants
Cell(Cambridge,Mass.), 133, 2008
|
|
4NIE
 
 | |
6XG6
 
 | | Full-length human mitochondrial Hsp90 (TRAP1) with ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Heat shock protein 75 kDa, ... | | Authors: | Liu, Y.X, Wang, F, Agard, D.A. | | Deposit date: | 2020-06-17 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | General and robust covalently linked graphene oxide affinity grids for high-resolution cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4DH6
 
 | | Structure of Bace-1 (Beta-Secretase) in Complex with (2R)-N-((2S,3R)-1-(benzo[d][1,3]dioxol-5-yl)-3-hydroxy-4-((S)-6'-neopentyl-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridine]-4'-ylamino)butan-2-yl)-2-methoxypropanamide | | Descriptor: | (2R)-N-[(2S,3R)-1-(1,3-benzodioxol-5-yl)-4-{[(4'S)-6'-(2,2-dimethylpropyl)-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl]amino}-3-hydroxybutan-2-yl]-2-methoxypropanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2012-01-27 | | Release date: | 2012-04-18 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and preparation of a potent series of hydroxyethylamine containing beta-secretase inhibitors that demonstrate robust reduction of central beta-amyloid.
J.Med.Chem., 55, 2012
|
|
4DI2
 
 | | Crystal structure of BACE1 in complex with hydroxyethylamine inhibitor 37 | | Descriptor: | (2R)-N-{(2S,3R)-4-{[(4'S)-6'-(2,2-dimethylpropyl)-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl]amino}-3-hydroxy-1-[3-(1,3-thiazol-2-yl)phenyl]butan-2-yl}-2-methoxypropanamide, Beta-secretase 1, GLYCEROL | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2012-01-30 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of potent, orally efficacious hydroxyethylamine derived beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors.
J.Med.Chem., 55, 2012
|
|
7LVA
 
 | | Solution structure of the HIV-1 PBS-segment | | Descriptor: | RNA (103-MER) | | Authors: | Heng, X, Song, Z. | | Deposit date: | 2021-02-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | The three-way junction structure of the HIV-1 PBS-segment binds host enzyme important for viral infectivity.
Nucleic Acids Res., 49, 2021
|
|
