2AOG
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog P2-NC | | Descriptor: | ACETIC ACID, GLYCEROL, HIV-1 PROTEASE (RETROPEPSIN), ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2F8G
 
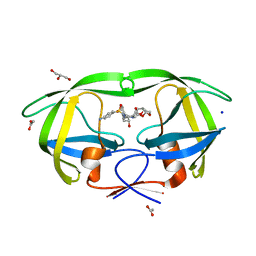 | | HIV-1 protease mutant I50V complexed with inhibitor TMC114 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2005-12-02 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M.
J.Med.Chem., 49, 2006
|
|
2F80
 
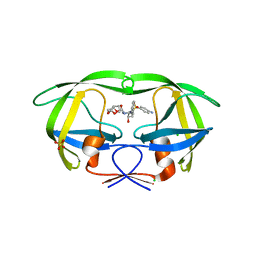 | | HIV-1 Protease mutant D30N complexed with inhibitor TMC114 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, POL POLYPROTEIN, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2005-12-01 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M.
J.Med.Chem., 49, 2006
|
|
2F81
 
 | | HIV-1 Protease mutant L90M complexed with inhibitor TMC114 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2005-12-01 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M.
J.Med.Chem., 49, 2006
|
|
6O54
 
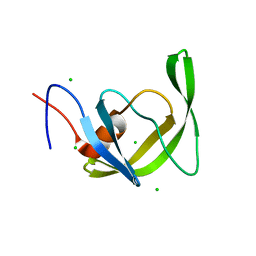 | | Crystal Structure of multi-drug resistant HIV-1 protease PR-S17 (D25N) | | Descriptor: | CHLORIDE ION, HIV-1 protease | | Authors: | Wang, Y.-F, Brothers, R, Agniswamy, J, Weber, I.T. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Highly Drug-Resistant HIV-1 Protease Mutant PRS17 Shows Enhanced Binding to Substrate Analogues.
Acs Omega, 4, 2019
|
|
6O5X
 
 | | Crystal Structure of multi-drug resistant HIV-1 protease PR-S17 with substrate analog CA-p2 | | Descriptor: | CHLORIDE ION, HIV-1 protease PR-S17, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, ... | | Authors: | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2019-03-04 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Highly Drug-Resistant HIV-1 Protease Mutant PRS17 Shows Enhanced Binding to Substrate Analogues.
Acs Omega, 4, 2019
|
|
6O5A
 
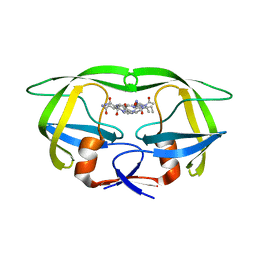 | |
6O48
 
 | | Wild-type HIV-1 protease in complex with a substrate analog CA-p2 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Liu, F, Weber, I.T. | | Deposit date: | 2019-02-28 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Highly Drug-Resistant HIV-1 Protease Mutant PRS17 Shows Enhanced Binding to Substrate Analogues.
Acs Omega, 4, 2019
|
|
6O57
 
 | |
6OTG
 
 | | HIV-1 protease triple mutants V32I, I47V, V82I with GRL-011-11A (a methylamine bis-Tetrahydrofuran P2-Ligand, sulfonamide isostere derivate) | | Descriptor: | (3R,3aS,4R,6aR)-4-(methylamino)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Pawar, S, Weber, I.T. | | Deposit date: | 2019-05-03 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural studies of antiviral inhibitor with HIV-1 protease bearing drug resistant substitutions of V32I, I47V and V82I.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
6PRF
 
 | | HIV-1 Protease multiple drug resistant clinical isolate mutant PR20 with GRL-14213A | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease, YTTRIUM ION | | Authors: | Kneller, D.W, Agniswamy, J, Weber, I.T. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor combats highly drug resistant mutant PR20.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
6P9B
 
 | |
6P9A
 
 | | HIV-1 Protease multiple mutant PRS5B with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 Protease, PHOSPHATE ION | | Authors: | Kneller, D.W, Agniswamy, J, Weber, I.T. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Highly drug-resistant HIV-1 protease reveals decreased intra-subunit interactions due to clusters of mutations.
Febs J., 287, 2020
|
|
2OGS
 
 | | Crystal Structure of the GEOBACILLUS STEAROTHERMOPHILUS Carboxylesterase EST55 at pH 6.2 | | Descriptor: | IODIDE ION, Thermostable carboxylesterase Est50 | | Authors: | Liu, P, Ewis, H.E, Tai, P.C, Lu, C.D, Weber, I.T. | | Deposit date: | 2007-01-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est55 and Its Activation of Prodrug CPT-11.
J.Mol.Biol., 367, 2007
|
|
2OGT
 
 | | Crystal Structure of the Geobacillus Stearothermophilus Carboxylesterase EST55 at pH 6.8 | | Descriptor: | GLYCEROL, IODIDE ION, Thermostable carboxylesterase Est50 | | Authors: | Liu, P, Ewis, H.E, Tai, P.C, Lu, C.D, Weber, I.T. | | Deposit date: | 2007-01-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est55 and Its Activation of Prodrug CPT-11.
J.Mol.Biol., 367, 2007
|
|
2NMZ
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEASE, SULFATE ION | | Authors: | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-10-23 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2NNK
 
 | | Crystal structure analysis of HIV-1 protease mutant I84V with a inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2NMY
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, PROTEASE, ... | | Authors: | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-10-23 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2NNP
 
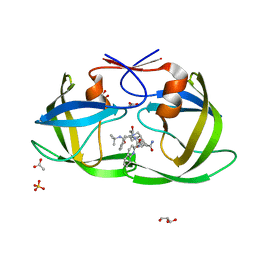 | | Crystal structure analysis of HIV-1 protease mutant I84V with a inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, GLYCEROL, ... | | Authors: | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2QCI
 
 | | HIV-1 Protease mutant D30N with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
2QD8
 
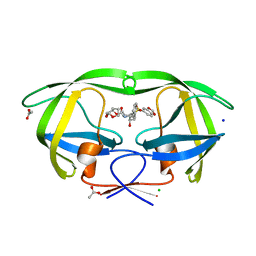 | | HIV-1 Protease Mutant I84V with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
2QD7
 
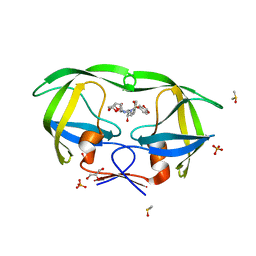 | | HIV-1 Protease Mutant V82A with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
3LJP
 
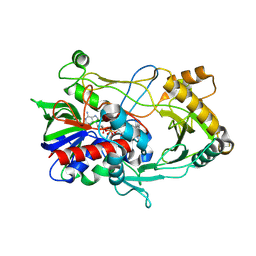 | | Crystal structure of choline oxidase V464A mutant | | Descriptor: | Choline oxidase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Finnegan, S, Agniswamy, J, Weber, I.T, Giovanni, G. | | Deposit date: | 2010-01-26 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of valine 464 in the flavin oxidation reaction catalyzed by choline oxidase.
Biochemistry, 49, 2010
|
|
3NU6
 
 | |
3NU5
 
 | | Crystal Structure of HIV-1 Protease Mutant I50V with Antiviral Drug Amprenavir | | Descriptor: | ACETATE ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Wang, Y.-F, Shen, C.H, Weber, I.T. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
