7W1S
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-007 | | Descriptor: | Nanobody Nb-007, Spike protein S1 | | Authors: | Yang, J, Lin, S, Sun, H.L, Lu, G.W. | | Deposit date: | 2021-11-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | A Potent Neutralizing Nanobody Targeting the Spike Receptor-Binding Domain of SARS-CoV-2 and the Structural Basis of Its Intimate Binding.
Front Immunol, 13, 2022
|
|
6JM5
 
 | | Crystal structure of TBC1D23 C terminal domain | | Descriptor: | SODIUM ION, TBC1 domain family member 23 | | Authors: | Sun, Q, Huang, W. | | Deposit date: | 2019-03-07 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional studies of TBC1D23 C-terminal domain provide a link between endosomal trafficking and PCH.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7XCH
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (two-RBD-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XA7
 
 | | Crystal structure of SARS-CoV-2 receptor-binding domain in complex with intermediate horseshoe bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Tang, L.F, Zhang, D, Han, P, Qi, J.X. | | Deposit date: | 2022-03-17 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis of SARS-CoV-2 and its variants binding to intermediate horseshoe bat ACE2.
Int J Biol Sci, 18, 2022
|
|
6OD3
 
 | | Human TCF4 C-terminal bHLH domain in Complex with 13-bp Oligonucleotide Containing E-box Sequence | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*AP*TP*AP*CP*AP*CP*GP*TP*GP*TP*AP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
7RCZ
 
 | | Crystal structure of C. difficile SpoVD in complex with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RD0
 
 | |
7RCY
 
 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein, ZINC ION | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RCX
 
 | |
7RCW
 
 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7YA0
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (S-6P-RRAR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
6OD5
 
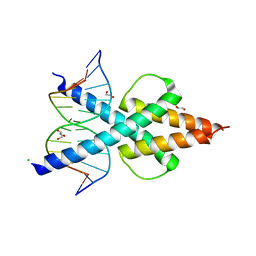 | | Human TCF4 C-terminal bHLH domain in Complex with 12-bp Oligonucleotide Containing E-box Sequence with 5-carboxylcytosines | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*AP*(1CC)P*GP*CP*AP*CP*GP*TP*GP*(1CC)P*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
6OD4
 
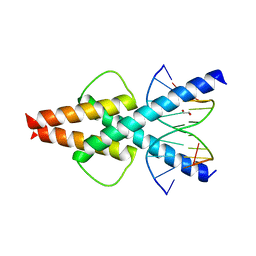 | | Human TCF4 C-terminal bHLH domain in Complex with 11-bp Oligonucleotide Containing E-box Sequence | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*AP*CP*AP*CP*GP*TP*GP*TP*A)-3'), Transcription factor 4 | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
7Y0Z
 
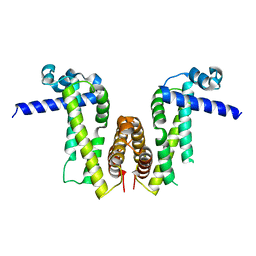 | | Crystal structure of Pseudomonas aeruginosa PvrA | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
7Y0Y
 
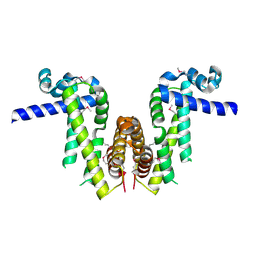 | | Crystal structure of Pseudomonas aeruginosa PvrA (SeMet) | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
6PBD
 
 | | DNA N6-Adenine Methyltransferase CcrM In Complex with Double-Stranded DNA Oligonucleotide Containing Its Recognition Sequence GAATC | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*AP*TP*TP*CP*AP*AP*TP*GP*AP*AP*TP*CP*CP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*TP*GP*GP*GP*AP*TP*TP*CP*AP*TP*TP*GP*AP*AP*TP*C)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Woodcock, C.B. | | Deposit date: | 2019-06-13 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | The cell cycle-regulated DNA adenine methyltransferase CcrM opens a bubble at its DNA recognition site.
Nat Commun, 10, 2019
|
|
7Y9S
 
 | | Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-26 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7FC0
 
 | | Reconstitution of MbnABC complex from Rugamonas rubra ATCC-43154 (GroupIII) | | Descriptor: | FE (III) ION, Methanobactin biosynthesis cassette protein MbnB, Methanobactin biosynthesis cassette protein MbnC, ... | | Authors: | Chao, D, Zhaolin, L, Shoujie, L, Li, Z, Dan, Z, Ying, J, Wei, C. | | Deposit date: | 2021-07-13 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.643 Å) | | Cite: | Crystal structure and catalytic mechanism of the MbnBC holoenzyme required for methanobactin biosynthesis.
Cell Res., 32, 2022
|
|
7BH2
 
 | | Cryo-EM Structure of KdpFABC in E2Pi state with BeF3 and K+ | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7WA9
 
 | |
7BPC
 
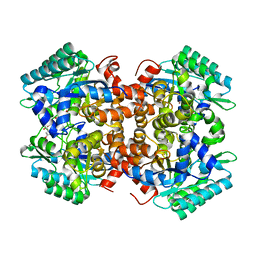 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with 2,5-DHBA | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, 2,5-dihydroxybenzoic acid, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-22 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
7BH1
 
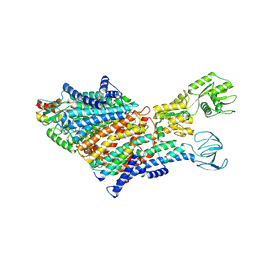 | | Cryo-EM Structure of KdpFABC in E1 state with K | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
