4MLN
 
 | | Crystal of PhnZ bound to (R)-2-amino-1-hydroxyethylphosphonic acid | | Descriptor: | FE (III) ION, Predicted HD phosphohydrolase PhnZ, [(1R)-2-amino-1-hydroxyethyl]phosphonic acid | | Authors: | van Staalduinen, L.M, McSorley, F.R, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PhnZ in complex with substrate reveals a di-iron oxygenase mechanism for catabolism of organophosphonates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P0U
 
 | |
4MGF
 
 | |
3P2U
 
 | | Crystal structure of PhnP in complex with orthovanadate | | Descriptor: | MANGANESE (II) ION, Protein phnP, VANADATE ION, ... | | Authors: | Podzelinska, K, Kim, Y.M, Zechel, D, Faucher, F, Jia, Z. | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of PhnP in complex with orthovanadate
TO BE PUBLISHED
|
|
6MSI
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 | | Descriptor: | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 | | Authors: | Deluca, C.I, Davies, P.L, Ye, Q, Jia, Z. | | Deposit date: | 1997-09-17 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The effects of steric mutations on the structure of type III antifreeze protein and its interaction with ice.
J.Mol.Biol., 275, 1998
|
|
3PDT
 
 | |
5FDN
 
 | |
6KJ7
 
 | | E. coli ATCase catalytic subunit mutant - G166P | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit | | Authors: | Lei, Z, Zheng, J, Jia, Z. | | Deposit date: | 2019-07-21 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.839 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
6IIH
 
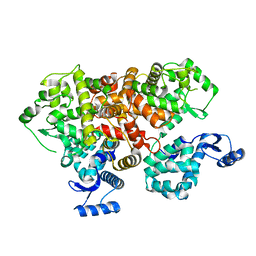 | | crystal structure of mitochondrial calcium uptake 2(MICU2) | | Descriptor: | CALCIUM ION, Endolysin,Calcium uptake protein 2, mitochondrial | | Authors: | Shen, Q, Wu, W, Zheng, J, Jia, Z. | | Deposit date: | 2018-10-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | The crystal structure of MICU2 provides insight into Ca2+binding and MICU1-MICU2 heterodimer formation.
Embo Rep., 20, 2019
|
|
6K5L
 
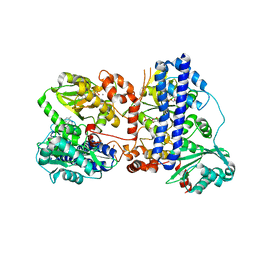 | | The crystal structure of isocitrate dehydrogenase kinase/phosphatase wtih two Mn2+ from E. coli | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Isocitrate dehydrogenase kinase/phosphatase, ... | | Authors: | Zhang, X, Lei, Z, Zheng, J, Jia, Z. | | Deposit date: | 2019-05-29 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characterization of metal binding of bifunctional kinase/phosphatase AceK and implication in activity modulation.
Sci Rep, 9, 2019
|
|
4WY8
 
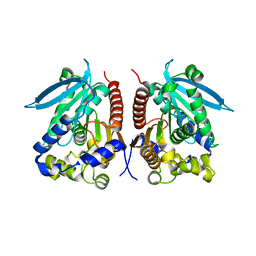 | | Structural analysis of two fungal esterases from Rhizomucor miehei explaining their substrate specificity | | Descriptor: | esterase | | Authors: | Qin, Z, Yang, S, Duan, X, Yan, Q, Jiang, Z. | | Deposit date: | 2014-11-16 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei
J.Lipid Res., 56, 2015
|
|
7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7CXM
 
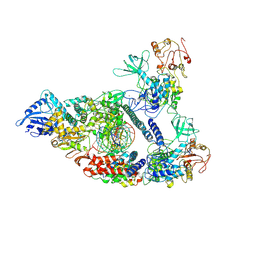 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
4WTR
 
 | | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaribiose | | Descriptor: | beta-1,3-glucanosyltransferase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | Deposit date: | 2014-10-30 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WY5
 
 | | Structural analysis of two fungal esterases from Rhizomucor miehei explaining their substrate specificity | | Descriptor: | Esterase, SULFATE ION | | Authors: | Qin, Z, Yang, S, Duan, X, Yan, Q, Jiang, Z. | | Deposit date: | 2014-11-15 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei
J.Lipid Res., 56, 2015
|
|
4WTP
 
 | | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei | | Descriptor: | 1,2-ETHANEDIOL, beta-1,3-glucanosyltransferase | | Authors: | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | Deposit date: | 2014-10-30 | | Release date: | 2015-08-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WTS
 
 | | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaritriose | | Descriptor: | beta-1,3-glucanosyltransferase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | Deposit date: | 2014-10-30 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5WDK
 
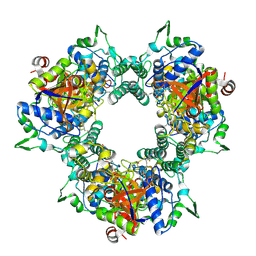 | | A processive dipeptidyl aminopeptidase secreted from an established commensal bacterium P. distasonis | | Descriptor: | 5-[(3aS,4R,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-N-(2-oxopropyl)pentanamide, Aminopeptidase C, POTASSIUM ION | | Authors: | Wolan, D.W, Xu, J.H, Solania, A, Chatterjee, S, Jiang, Z, ODonoghue, A.J. | | Deposit date: | 2017-07-05 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A Commensal Dipeptidyl Aminopeptidase with Specificity for N-Terminal Glycine Degrades Human-Produced Antimicrobial Peptides in Vitro.
Acs Chem.Biol., 13, 2018
|
|
5WDL
 
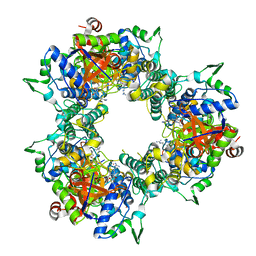 | | A processive dipeptidyl aminopeptidase secreted from an established commensal bacterium P. distasonis | | Descriptor: | Aminopeptidase C, N-[(3S)-6-carbamimidamido-2-oxohexan-3-yl]glycinamide, POTASSIUM ION | | Authors: | Wolan, D.W, Xu, J.H, Solania, A, Chatterjee, S, Jiang, Z, ODonoghue, A.J. | | Deposit date: | 2017-07-05 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.625 Å) | | Cite: | A Commensal Dipeptidyl Aminopeptidase with Specificity for N-Terminal Glycine Degrades Human-Produced Antimicrobial Peptides in Vitro.
Acs Chem.Biol., 13, 2018
|
|
4ZM6
 
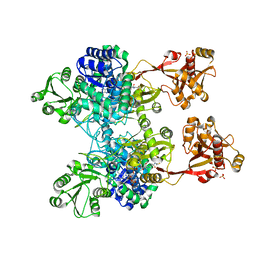 | | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase | | Descriptor: | ACETYL COENZYME *A, N-acetyl-beta-D glucosaminidase, SULFATE ION | | Authors: | Qin, Z, Xiao, Y, Yang, X, Jiang, Z, Yang, S, Mesters, J.R. | | Deposit date: | 2015-05-02 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain glycoside hydrolase family 3 beta-N-acetylglucosaminidase
Sci Rep, 5, 2015
|
|
3DSH
 
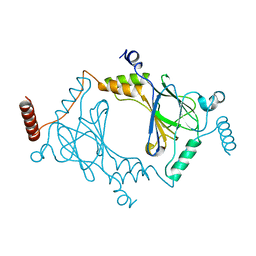 | | Crystal structure of dimeric interferon regulatory factor 5 (IRF-5) transactivation domain | | Descriptor: | Interferon regulatory factor 5 | | Authors: | Chen, W, Lam, S.S, Srinath, H, Jiang, Z, Correia, J.J, Schiffer, C, Fitzgerald, K.A, Lin, K, Royer Jr, W.E. | | Deposit date: | 2008-07-12 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5JVV
 
 | | Crystal structure and characterization an elongating GH family 16 beta-1,3-glucosyltransferase | | Descriptor: | beta-1,3-glucosyltransferase | | Authors: | Qin, Z, Yan, Q, Yang, S, Jiang, Z. | | Deposit date: | 2016-05-11 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Catalytic Mechanism of a Novel Glycoside Hydrolase Family 16 "Elongating" beta-Transglycosylase
J. Biol. Chem., 292, 2017
|
|
3WNV
 
 | | Crystal structure of a glyoxylate reductase from Paecilomyes thermophila | | Descriptor: | SULFATE ION, glyoxylate reductase | | Authors: | Duan, X, Hu, S, Zhou, P, Zhou, Y, Jiang, Z. | | Deposit date: | 2013-12-17 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and crystal structure of a first fungal glyoxylate reductase from Paecilomyes thermophila
Enzyme.Microb.Technol., 60, 2014
|
|
5H9X
 
 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii | | Descriptor: | beta-1,3-glucanase | | Authors: | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | Deposit date: | 2015-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
5H9Y
 
 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii complexed with laminarihexaose. | | Descriptor: | L(+)-TARTARIC ACID, beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | Deposit date: | 2015-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
