7XMZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D2 heavy chain, D2 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
7XMX
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three F61 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F61 heavy chain, F61 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
5V8O
 
 | | Discovery of a high affinity inhibitor of cGAS | | Descriptor: | 5-phenyltetrazolo[1,5-a]pyrimidin-7-ol, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Hall, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay.
PLoS ONE, 12, 2017
|
|
7EAM
 
 | | immune complex of SARS-CoV-2 RBD and cross-neutralizing antibody 7D6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Li, T.T, Gu, Y, Li, S.W. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants.
Nat Commun, 12, 2021
|
|
8XJ4
 
 | | Structure of prostatic acid phosphatase in human semen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prostatic acid phosphatase, alpha-D-mannopyranose, ... | | Authors: | Liu, X.Z, Li, J.L, Deng, D, Wang, X. | | Deposit date: | 2023-12-20 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Purification, identification and Cryo-EM structure of prostatic acid phosphatase in human semen.
Biochem.Biophys.Res.Commun., 702, 2024
|
|
7EAX
 
 | | Crystal complex of p53-V272M and antimony ion | | Descriptor: | ANTIMONY (III) ION, Cellular tumor antigen p53, ZINC ION | | Authors: | Lu, M, Tang, Y. | | Deposit date: | 2021-03-08 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Repurposing antiparasitic antimonials to noncovalently rescue temperature-sensitive p53 mutations.
Cell Rep, 39, 2022
|
|
8HVS
 
 | |
3G86
 
 | | Hepatitis C virus polymerase NS5B (BK 1-570) with thiazine inhibitor | | Descriptor: | N-{3-[6-fluoro-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl]-1,1-dioxido-4H-1,4-benzothiazin-7-yl}methanesulfonamide, NICKEL (II) ION, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-02-11 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 2: Synthesis and structure-activity relationships of benzothiazine-substituted quinolinediones
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7XST
 
 | |
2F1G
 
 | | Cathepsin S in complex with non-covalent 2-(Benzoxazol-2-ylamino)-acetamide | | Descriptor: | Cathepsin S, GLYCEROL, N~2~-1,3-BENZOXAZOL-2-YL-3-CYCLOHEXYL-N-{2-[(4-METHOXYPHENYL)AMINO]ETHYL}-L-ALANINAMIDE | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2005-11-14 | | Release date: | 2006-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and evaluation of arylaminoethyl amides as noncovalent inhibitors of cathepsin S. Part 3: Heterocyclic P3.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
7V97
 
 | | Arsenic-bound p53 DNA-binding domain mutant V272M | | Descriptor: | ARSENIC, Cellular tumor antigen p53, ZINC ION | | Authors: | Lu, M, Xing, Y.F, Wang, Z.Y, Ni, Y, Song, H.X. | | Deposit date: | 2021-08-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Diverse rescue potencies of p53 mutations to ATO are predetermined by intrinsic mutational properties.
Sci Transl Med, 15, 2023
|
|
5TPU
 
 | | x-ray structure of the WlaRB TDP-quinovose 3,4-ketoisomerase from campylobacter jejuni | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Holden, H.M, Thoden, J.B, Li, J.Z, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinogradov, E, Schoenhofen, I.C, Gilbert, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
7KEO
 
 | | Crystal structure of K29-linked di-ubiquitin in complex with synthetic antigen binding fragment | | Descriptor: | PHOSPHATE ION, Synthetic antigen binding fragment, heavy chain, ... | | Authors: | Yu, Y, Zheng, Q, Erramilli, S, Pan, M, Kossiakoff, A, Liu, L, Zhao, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle.
Nat.Chem.Biol., 17, 2021
|
|
7DV4
 
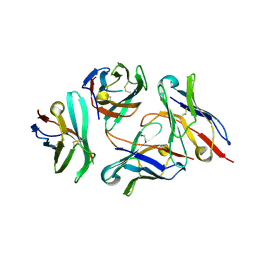 | | Crystal structure of anti-CTLA-4 VH domain in complex with human CTLA-4 | | Descriptor: | 1,2-ETHANEDIOL, 4003-1(VH), Cytotoxic T-lymphocyte protein 4 | | Authors: | Li, H, Gan, X, He, Y. | | Deposit date: | 2021-01-12 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | An anti-CTLA-4 heavy chain-only antibody with enhanced T reg depletion shows excellent preclinical efficacy and safety profile.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8J6L
 
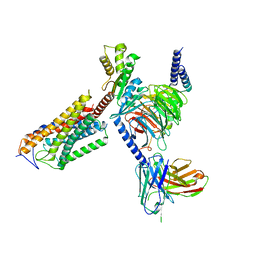 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound niacin | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
8J6I
 
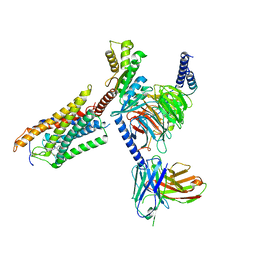 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound MK-6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
8JPC
 
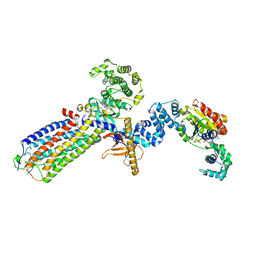 | | cryo-EM structure of NTSR1-GRK2-Galpha(q) complexes 2 | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, Beta-adrenergic receptor kinase 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Duan, J, Liu, H, Zhao, F, Yuan, Q, Ji, Y, Xu, H.E. | | Deposit date: | 2023-06-11 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | GPCR activation and GRK2 assembly by a biased intracellular agonist.
Nature, 620, 2023
|
|
8JPF
 
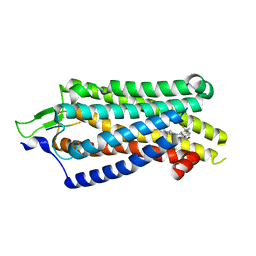 | | Focused refiment structure of NTSR1 in NTSR1-GRK2-Galpha(q) complexes | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, NTS, Neurotensin receptor type 1 | | Authors: | Duan, J, Liu, H, Zhao, F, Yuan, Q, Ji, Y, Xu, H.E. | | Deposit date: | 2023-06-11 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | GPCR activation and GRK2 assembly by a biased intracellular agonist.
Nature, 620, 2023
|
|
8JPD
 
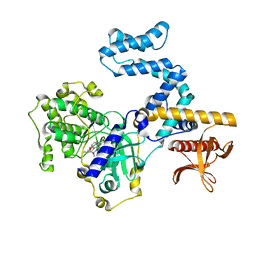 | | Focused refinement structure of GRK2 in NTSR1-GRK2-Galpha(q) complexes | | Descriptor: | Beta-adrenergic receptor kinase 1, STAUROSPORINE | | Authors: | Duan, J, Liu, H, Zhao, F, Yuan, Q, Ji, Y, Xu, H.E. | | Deposit date: | 2023-06-11 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | GPCR activation and GRK2 assembly by a biased intracellular agonist.
Nature, 620, 2023
|
|
8JPE
 
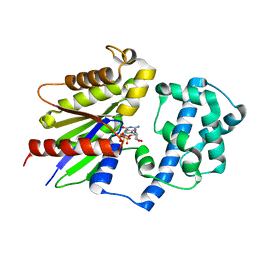 | | Focused refinement structure of Galpha(q) in NTSR1-GRK2-Galpha(q) complexes | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(q) subunit alpha, MAGNESIUM ION, ... | | Authors: | Duan, J, Liu, H, Zhao, F, Yuan, Q, Ji, Y, Xu, H.E. | | Deposit date: | 2023-06-11 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | GPCR activation and GRK2 assembly by a biased intracellular agonist.
Nature, 620, 2023
|
|
7JOU
 
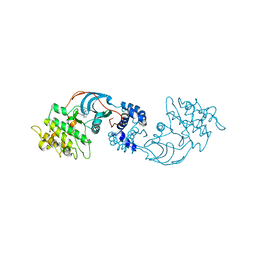 | |
8WRJ
 
 | | glycosyltransferase UGT74AN3 | | Descriptor: | 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Huang, W, Long, F. | | Deposit date: | 2023-10-15 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8WRK
 
 | | glycosyltransferase UGT74AN3 | | Descriptor: | Glycosyltransferase, PICEATANNOL, URIDINE-5'-DIPHOSPHATE | | Authors: | Huang, W, Long, F. | | Deposit date: | 2023-10-15 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8J6J
 
 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound with GSK256073 | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
7JOV
 
 | |
