8CE3
 
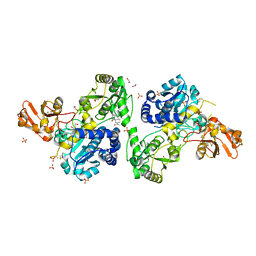 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with 3D fragment 2548 | | Descriptor: | (3~{S})-1-ethanoyl-3-(4-methylphenyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Gilio, A.K, Darby, J.F, Wu, L, Davies, G.J. | | Deposit date: | 2023-02-01 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Modular 3-D Fragment Collection: Design, Synthesis and Screening
To Be Published
|
|
4BY9
 
 | | The structure of the Box CD enzyme reveals regulation of rRNA methylation | | Descriptor: | 5'-R(*UP*CP*GP*CP*CP*CP*AP*UP*CP*AP*CP)-3', 50S RIBOSOMAL PROTEIN L7AE, FIBRILLARIN-LIKE RRNA/TRNA 2'-O-METHYLTRANSFERASE, ... | | Authors: | Lapinaite, A, Simon, B, Skjaerven, L, Rakwalska-Bange, M, Gabel, F, Carlomagno, T. | | Deposit date: | 2013-07-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Box C/D Enzyme Reveals Regulation of RNA Methylation.
Nature, 502, 2013
|
|
8CD6
 
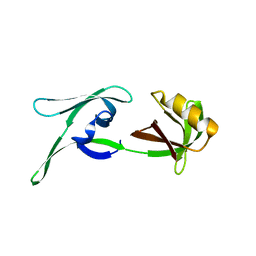 | | structure of HEX-1 (cyto V2) from N. crassa grown in living insect cells, diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
6GP1
 
 | | Structure of mEos4b in the red long-lived dark state | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP | | Authors: | De Zitter, E, Adam, V, Byrdin, M, Van Meervelt, L, Dedecker, P, Bourgeois, D. | | Deposit date: | 2018-06-04 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Mechanistic investigation of mEos4b reveals a strategy to reduce track interruptions in sptPALM.
Nat.Methods, 16, 2019
|
|
8CK2
 
 | | MUC2 CysD1 G1352S | | Descriptor: | ACETATE ION, CALCIUM ION, Mucin-2, ... | | Authors: | Khmelnitsky, L, Fass, D. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pulmonary Fibrosis Mutation Undermines Mucin 5B Supramolecular Assembly
To Be Published
|
|
1CW9
 
 | | DNA DECAMER WITH AN ENGINEERED CROSSLINK IN THE MINOR GROOVE | | Descriptor: | 5'-D(*CP*CP*AP*GP*(G47)P*CP*CP*TP*GP*G)-3', CALCIUM ION | | Authors: | van Aalten, D.M.F, Erlanson, D.A, Verdine, G.L, Joshua-Tor, L. | | Deposit date: | 1999-08-26 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A structural snapshot of base-pair opening in DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
8CDC
 
 | | Native 3CLpro from SARS-CoV-2 at 1.54 A | | Descriptor: | 3C-like proteinase | | Authors: | Mazzei, L, Jovanovic, A, Acton, T.B, Ciurli, S, Montelione, G.T. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Expression, purification, and characterization of SARS-CoV2 3CLpro in a mature form
To Be Published
|
|
5X72
 
 | | The crystal Structure PDE delta in complex with (rac)-p9 | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, (2S)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
1O7E
 
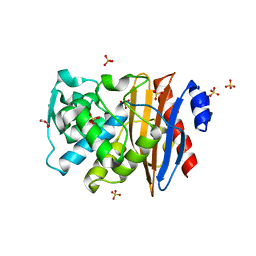 | |
5XJ0
 
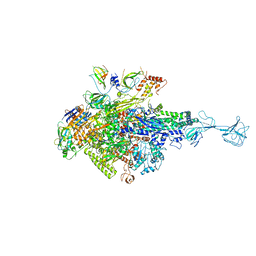 | | T. thermophilus RNA polymerase holoenzyme bound with gp39 and gp76 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Ooi, W.Y, Murayama, Y, Mekler, V, Minakhin, L, Severinov, K, Yokoyama, S, Sekine, S. | | Deposit date: | 2017-04-28 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | A Thermus phage protein inhibits host RNA polymerase by preventing template DNA strand loading during open promoter complex formation
Nucleic Acids Res., 46, 2018
|
|
5MF9
 
 | | Solution structure of the RBM5 OCRE domain in complex with polyproline SmN peptide. | | Descriptor: | RNA-binding protein 5, Survival motor neuron protein | | Authors: | Mourao, A, Sattler, M, Bonnal, S, Komal, S, Warner, L, Bordonne, R, Valcarcel, J. | | Deposit date: | 2016-11-17 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition ofspliceosomal SmN B B proteins by theRBM5 OCRE domain in splicing regulation
Elife, 5, 2016
|
|
8RXR
 
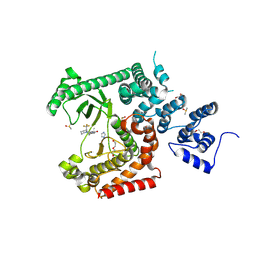 | | Crystal structure of VPS34 in complex with inhibitor SB02024 | | Descriptor: | 4-[(3R)-3-methylmorpholin-4-yl]-2-[(2R)-2-(trifluoromethyl)piperidin-1-yl]-3H-pyridin-6-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Tresaugues, L, Yu, Y, Bogdan, M, Parpal, S, Silvander, C, Lindstrom, J, Simeon, J, Timson, M.J, Al-Hashimi, H, Smith, B.D, Flynn, D.L, Viklund, J, Martinsson, J, De Milito, A, Andersson, M. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Combining VPS34 inhibitors with STING agonists enhances type I interferon signaling and anti-tumor efficacy.
Mol Oncol, 18, 2024
|
|
3V70
 
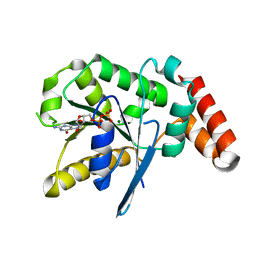 | | Crystal Structure of Human GTPase IMAP family member 1 | | Descriptor: | GTPase IMAP family member 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Nedyalkova, L, Shen, Y, Tong, Y, Tempel, W, Mackenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Andrews, D.W, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Crystal Structure of Human GTPase IMAP family member 1
to be published
|
|
6GJW
 
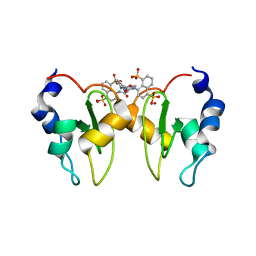 | | Structure of XIAP-BIR1 domain in complex with an NF023 analog | | Descriptor: | 4-[[3-[[3-[(4,8-disulfonatonaphthalen-1-yl)carbamoyl]phenyl]carbamoylamino]phenyl]carbonylamino]naphthalene-1,5-disulfonate, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Sorrentino, L, Cossu, F, Malkoc, B, Zaffaroni, M, Milani, M, Mastrangelo, E. | | Deposit date: | 2018-05-17 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Relationship of NF023 Derivatives Binding to XIAP-BIR1.
Chemistryopen, 8, 2019
|
|
8WOU
 
 | |
4BP1
 
 | | Crystal Structure of Plasmodium Falciparum Spermidine Synthase in Complex with 5'-Methylthioadenosine and Putrescine | | Descriptor: | 1,4-DIAMINOBUTANE, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Sprenger, J, Halander, J.C, Svensson, B, Al-Karadaghi, S, Persson, L. | | Deposit date: | 2013-05-23 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Three-Dimensional Structures of Plasmodium Falciparum Spermidine Synthase with Bound Inhibitors Suggest New Strategies for Drug Design
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1D5W
 
 | | PHOSPHORYLATED FIXJ RECEIVER DOMAIN | | Descriptor: | SULFATE ION, TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Birck, C, Mourey, L, Gouet, P, Fabry, B, Schumacher, J, Rousseau, P, Kahn, D, Samama, J.P. | | Deposit date: | 1999-10-12 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational changes induced by phosphorylation of the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
8WMK
 
 | | Structure of ConA/Man2 | | Descriptor: | 4-[3-(4-carboxyphenyl)phenyl]benzoic acid, CALCIUM ION, Concanavalin-A, ... | | Authors: | Li, L, Chen, G. | | Deposit date: | 2023-10-03 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Rational design of inducing ligands with three-dimensional supramolecular interactions to build protein crystalline frameworks.
To Be Published
|
|
3UN6
 
 | | 2.0 Angstrom Crystal Structure of Ligand Binding Component of ABC-type Import System from Staphylococcus aureus with Zinc bound | | Descriptor: | ABC transporter substrate-binding protein, PHOSPHATE ION, ZINC ION | | Authors: | Minasov, G, Wawrzak, Z, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-15 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 2.0 Angstrom Crystal Structure of Ligand Binding Component of ABC-type Import System from Staphylococcus aureus with Zinc bound.
TO BE PUBLISHED
|
|
5XGV
 
 | |
8CO5
 
 | |
8WMG
 
 | | Structure of ConA/Man3 | | Descriptor: | 4-[3,5-bis(4-carboxyphenyl)phenyl]benzoic acid, CALCIUM ION, Concanavalin-A, ... | | Authors: | Li, L, Chen, G. | | Deposit date: | 2023-10-03 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Rational design of inducing ligands with three-dimensional supramolecular interactions to build protein crystalline frameworks.
To Be Published
|
|
8WKJ
 
 | |
1CF4
 
 | | CDC42/ACK GTPASE-BINDING DOMAIN COMPLEX | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN (ACTIVATED P21CDC42HS KINASE), ... | | Authors: | Mott, H.R, Owen, D, Nietlispach, D, Lowe, P.N, Lim, L, Laue, E.D. | | Deposit date: | 1999-03-23 | | Release date: | 1999-06-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK.
Nature, 399, 1999
|
|
4C04
 
 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with inhibitor | | Descriptor: | PROTEIN ARGININE N-METHYLTRANSFERASE 6, SINEFUNGIN | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
