4KGD
 
 | |
4KXX
 
 | | Human transketolase in covalent complex with donor ketose D-sedoheptulose-7-phosphate | | Descriptor: | (2R,3R,4S,5R,6S)-2,3,4,5,6,7-hexahydroxyheptyl dihydrogen phosphate, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
4KXU
 
 | | Human transketolase in covalent complex with donor ketose D-fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, D-SORBITOL-6-PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
4JQ9
 
 | | Dihydrolipoyl dehydrogenase of Escherichia coli pyruvate dehydrogenase complex | | Descriptor: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tietzel, M, Neumann, P, Meyer, D, Ficner, R, Tittmann, K. | | Deposit date: | 2013-03-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Dihydrolipoyl dehydrogenase of Escherichia coli pyruvate dehydrogenase complex
TO BE PUBLISHED
|
|
1DIP
 
 | | THE SOLUTION STRUCTURE OF PORCINE DELTA-SLEEP-INDUCING PEPTIDE IMMUNOREACTIVE PEPTIDE, NMR, 10 STRUCTURES | | Descriptor: | DELTA-SLEEP-INDUCING PEPTIDE IMMUNOREACTIVE PEPTIDE | | Authors: | Roesch, P, Seidel, G, Adermann, K, Schindler, T, Ejchart, A, Jaenicke, R, Forssmann, W.G. | | Deposit date: | 1997-04-09 | | Release date: | 1997-10-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine delta sleep-inducing peptide immunoreactive peptide A homolog of the shortsighted gene product.
J.Biol.Chem., 272, 1997
|
|
6Y4D
 
 | | Crystal structure of a short-chain dehydrogenase/reductase (SDR) from Zephyranthes treatiae in complex with NADP+ | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, short-chain dehydrogenase/reductase (SDR) | | Authors: | Sautner, V, Steimle, S, Roth, S, Mueller, M, Tittmann, K. | | Deposit date: | 2020-02-20 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crossing the Border: From Keto- to Imine Reduction in Short-Chain Dehydrogenases/Reductases.
Chembiochem, 21, 2020
|
|
6YJY
 
 | | Crystal structure of human glutaminyl cyclase in complex with neurotensin 1-5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTAMINE, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Funk, L.M, Sautner, V, Tittmann, K. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Hydrazides Are Potent Transition-State Analogues for Glutaminyl Cyclase Implicated in the Pathogenesis of Alzheimer's Disease.
Biochemistry, 59, 2020
|
|
6YI1
 
 | | Crystal structure of human glutaminyl cyclase in complex with Glu(gamma-hydrazide)-Phe-Ala | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kupski, O, Sautner, V, Tittmann, K. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Hydrazides Are Potent Transition-State Analogues for Glutaminyl Cyclase Implicated in the Pathogenesis of Alzheimer's Disease.
Biochemistry, 59, 2020
|
|
3IO1
 
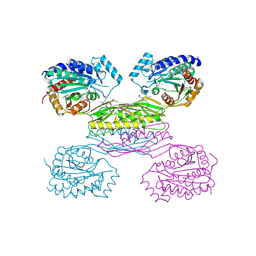 | | Crystal Structure of Aminobenzoyl-glutamate utilization protein from Klebsiella pneumoniae | | Descriptor: | Aminobenzoyl-glutamate utilization protein, SODIUM ION, YTTRIUM (III) ION | | Authors: | Kumaran, D, Baumann, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-13 | | Release date: | 2009-08-25 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Aminobenzoyl-glutamate utilization protein from Klebsiella pneumoniae
To be Published
|
|
6YR3
 
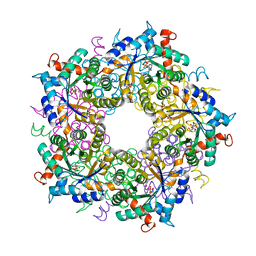 | |
6YRH
 
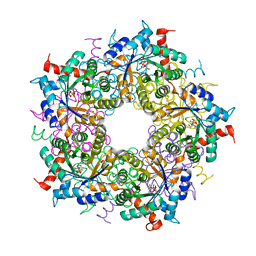 | |
6YRT
 
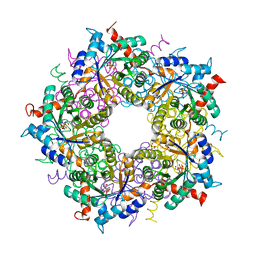 | |
6YS0
 
 | |
6YRE
 
 | | Transaldolase variant T30C/D211C from T. acidophilum | | Descriptor: | ACETATE ION, GLYCEROL, Probable transaldolase | | Authors: | Sautner, V, Lietzow, T.H, Tittmann, K. | | Deposit date: | 2020-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Large-scale motions underlie physical but not chemical steps in transaldolase mechanism: Substrate binding by conformational selection and rate-determining product release
To Be Published
|
|
6YRM
 
 | |
6QL5
 
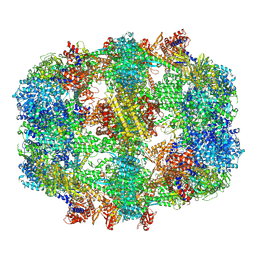 | | Structure of fatty acid synthase complex with bound gamma subunit from Saccharomyces cerevisiae at 2.8 angstrom | | Descriptor: | 4'-PHOSPHOPANTETHEINE, FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Graf, B, Linden, A, Sautner, V, Urlaub, H, Tittmann, K, Stark, H, Chari, A. | | Deposit date: | 2019-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase.
Cell, 180, 2020
|
|
6QL7
 
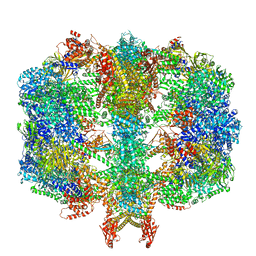 | | Structure of fatty acid synthase complex with bound gamma subunit from Saccharomyces cerevisiae at 4.6 angstrom | | Descriptor: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, Translation machinery-associated protein 17 | | Authors: | Singh, K, Graf, B, Linden, A, Sautner, V, Urlaub, H, Tittmann, K, Stark, H, Chari, A. | | Deposit date: | 2019-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase.
Cell, 180, 2020
|
|
6QL9
 
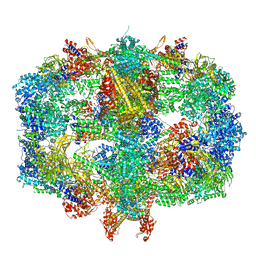 | | Structure of Fatty acid synthase complex from Saccharomyces cerevisiae at 2.9 Angstrom | | Descriptor: | 1,2-ETHANEDIOL, 4'-PHOSPHOPANTETHEINE, ACETATE ION, ... | | Authors: | Singh, K, Graf, B, Linden, A, Sautner, V, Urlaub, H, Tittmann, K, Stark, H, Chari, A. | | Deposit date: | 2019-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase.
Cell, 180, 2020
|
|
6QL6
 
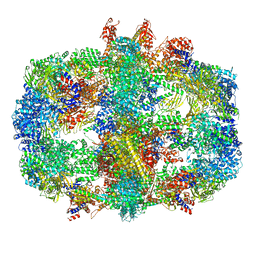 | | Structure of Fatty acid synthase complex from Saccharomyces cerevisiae at 2.9 Angstrom | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, ... | | Authors: | Singh, K, Graf, B, Linden, A, Sautner, V, Urlaub, H, Tittmann, K, Stark, H, Chari, A. | | Deposit date: | 2019-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase.
Cell, 180, 2020
|
|
1UYA
 
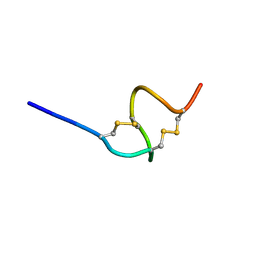 | | THE SOLUTION STRUCTURE OF THE A-FORM OF UROGUANYLIN-16 NMR, 10 STRUCTURES | | Descriptor: | UROGUANYLIN-16, ISOMER A | | Authors: | Marx, U.C, Adermann, K, Forssmann, W.-G, Roesch, P. | | Deposit date: | 1997-09-11 | | Release date: | 1998-03-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | One peptide, two topologies: structure and interconversion dynamics of human uroguanylin isomers.
J.Pept.Res., 52, 1998
|
|
1UYB
 
 | | THE SOLUTION STRUCTURE OF THE B-FORM OF UROGUANYLIN-16 NMR, 10 STRUCTURES | | Descriptor: | UROGUANYLIN-16, ISOMER B | | Authors: | Marx, U.C, Adermann, K, Forssmann, W.-G, Roesch, P. | | Deposit date: | 1997-09-11 | | Release date: | 1998-03-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | One peptide, two topologies: structure and interconversion dynamics of human uroguanylin isomers.
J.Pept.Res., 52, 1998
|
|
1HPY
 
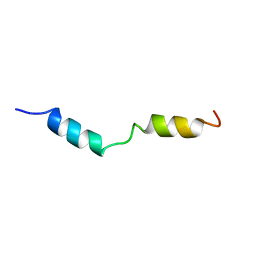 | | THE SOLUTION STRUCTURE OF HUMAN PARATHYROID HORMONE FRAGMENT 1-34 IN 20% TRIFLUORETHANOL, NMR, 10 STRUCTURES | | Descriptor: | PARATHYROID HORMONE | | Authors: | Marx, U.C, Roesch, P, Adermann, K, Bayer, P, Forssmann, W.-G. | | Deposit date: | 1998-09-30 | | Release date: | 2000-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human parathyroid hormone fragments hPTH(1-34) and hPTH(1-39) and bovine parathyroid hormone fragment bPTH(1-37).
Biochem.Biophys.Res.Commun., 267, 2000
|
|
6RJC
 
 | |
6RJB
 
 | |
8PZ3
 
 | |
